PIM serine/threonine kinases in the pathogenesis and therapy of hematologic malignancies and solid cancers
- PMID: 20145274
- PMCID: PMC2878801
- DOI: 10.3324/haematol.2009.017079
PIM serine/threonine kinases in the pathogenesis and therapy of hematologic malignancies and solid cancers
Abstract
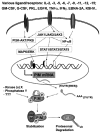
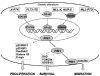
The identification as cooperating targets of Proviral Integrations of Moloney virus in murine lymphomas suggested early on that PIM serine/threonine kinases play an important role in cancer biology. Whereas elevated levels of PIM1 and PIM2 were mostly found in hematologic malignancies and prostate cancer, increased PIM3 expression was observed in different solid tumors. PIM kinases are constitutively active and their activity supports in vitro and in vivo tumor cell growth and survival through modification of an increasing number of common as well as isoform-specific substrates including several cell cycle regulators and apoptosis mediators. PIM1 but not PIM2 seems also to mediate homing and migration of normal and malignant hematopoietic cells by regulating chemokine receptor surface expression. Knockdown experiments by RNA interference or dominant-negative acting mutants suggested that PIM kinases are important for maintenance of a transformed phenotype and therefore potential therapeutic targets. Determination of the protein structure facilitated identification of an increasing number of potent small molecule PIM kinase inhibitors with in vitro and in vivo anticancer activity. Ongoing efforts aim to identify isoform-specific PIM inhibitors that would not only help to dissect the kinase function but hopefully also provide targeted therapeutics. Here, we summarize the current knowledge about the role of PIM serine/threonine kinases for the pathogenesis and therapy of hematologic malignancies and solid cancers, and we highlight structural principles and recent progress on small molecule PIM kinase inhibitors that are on their way into first clinical trials.
Figures



References
-
- Cuypers HT, Selten G, Quint W, Zijlstra M, Maandag ER, Boelens W, et al. Murine leukemia virus-induced T-cell lymphoma-genesis: integration of proviruses in a distinct chromosomal region. Cell. 1984;37(1):141–50. - PubMed
-
- Selten G, Cuypers HT, Boelens W, Robanus-Maandag E, Verbeek J, Domen J, et al. The primary structure of the putative oncogene pim-1 shows extensive homology with protein kinases. Cell. 1986;46(4):603–11. - PubMed
-
- van Lohuizen M, Verbeek S, Krimpenfort P, Domen J, Saris C, Radaszkiewicz T, et al. Predisposition to lymphomagenesis in pim-1 transgenic mice: cooperation with c-myc and N-myc in murine leukemia virus-induced tumors. Cell. 1989;56(4):673–82. - PubMed
-
- Berns A, Breuer M, Verbeek S, van Lohuizen M. Transgenic mice as a means to study synergism between oncogenes. Int J Cancer Suppl. 1989;4:22–5. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

