Simulation of DNA catenanes
- PMID: 20145800
- PMCID: PMC2845312
- DOI: 10.1039/b910812b
Simulation of DNA catenanes
Abstract
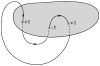
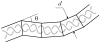
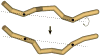
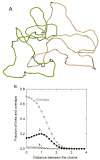
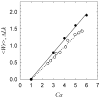
DNA catenanes are important objects in biology, foremost as they appear during replication of circular DNA molecules. In this review we analyze how conformational properties of DNA catenanes can be studied by computer simulation. We consider classification of catenanes, their topological invariants and the methods of calculation of these invariants. We briefly analyze the DNA model and the simulation procedure used to sample the equilibrium conformational ensemble of catenanes with a particular topology. We consider how to avoid direct simulation of many DNA molecules when we need to account for the linking-unlinking process. The simulation methods and their comparisons with experiments are illustrated by some examples. We also describe an approach that allows simulating the steady state fraction of DNA catenanes created by type II topoisomerases.
Figures
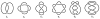




Similar articles
-
Unlinking of supercoiled DNA catenanes by type IIA topoisomerases.Biophys J. 2011 Sep 21;101(6):1403-11. doi: 10.1016/j.bpj.2011.08.011. Epub 2011 Sep 20. Biophys J. 2011. PMID: 21943421 Free PMC article.
-
Closing the DNA replication cycle: from simple circular molecules to supercoiled and knotted DNA catenanes.Nucleic Acids Res. 2019 Aug 22;47(14):7182-7198. doi: 10.1093/nar/gkz586. Nucleic Acids Res. 2019. PMID: 31276584 Free PMC article. Review.
-
DNA Catenation Reveals the Dynamics of DNA Topology During Replication.Methods Mol Biol. 2018;1703:75-86. doi: 10.1007/978-1-4939-7459-7_5. Methods Mol Biol. 2018. PMID: 29177734
-
DNA supercoiling and its role in DNA decatenation and unknotting.Nucleic Acids Res. 2010 Apr;38(7):2119-33. doi: 10.1093/nar/gkp1161. Epub 2009 Dec 21. Nucleic Acids Res. 2010. PMID: 20026582 Free PMC article. Review.
-
Molecular Dynamics Simulation of Supercoiled, Knotted, and Catenated DNA Molecules, Including Modeling of Action of DNA Gyrase.Methods Mol Biol. 2017;1624:339-372. doi: 10.1007/978-1-4939-7098-8_24. Methods Mol Biol. 2017. PMID: 28842894
Cited by
-
Unlinking of supercoiled DNA catenanes by type IIA topoisomerases.Biophys J. 2011 Sep 21;101(6):1403-11. doi: 10.1016/j.bpj.2011.08.011. Epub 2011 Sep 20. Biophys J. 2011. PMID: 21943421 Free PMC article.
-
Closing the DNA replication cycle: from simple circular molecules to supercoiled and knotted DNA catenanes.Nucleic Acids Res. 2019 Aug 22;47(14):7182-7198. doi: 10.1093/nar/gkz586. Nucleic Acids Res. 2019. PMID: 31276584 Free PMC article. Review.
-
Orientation of DNA Minicircles Balances Density and Topological Complexity in Kinetoplast DNA.PLoS One. 2015 Jun 25;10(6):e0130998. doi: 10.1371/journal.pone.0130998. eCollection 2015. PLoS One. 2015. PMID: 26110537 Free PMC article.
-
Pairing nanoarchitectonics of oligodeoxyribonucleotides with complex diversity: concatemers and self-limited complexes.RSC Adv. 2022 Feb 23;12(11):6416-6431. doi: 10.1039/d2ra00155a. eCollection 2022 Feb 22. RSC Adv. 2022. PMID: 35424594 Free PMC article.
-
Efficient deformation algorithm for plasmid DNA simulations.BMC Bioinformatics. 2014 Sep 15;15(1):301. doi: 10.1186/1471-2105-15-301. BMC Bioinformatics. 2014. PMID: 25225011 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

