Mesoscale simulations of two nucleosome-repeat length oligonucleosomes
- PMID: 20145817
- PMCID: PMC3182521
- DOI: 10.1039/b918629h
Mesoscale simulations of two nucleosome-repeat length oligonucleosomes
Abstract
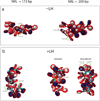
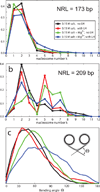
The compaction of chromatin, accessed through coarse-grained modeling and simulation, reveals different folding patterns as a function of the nucleosome repeat length (NRL), the presence of the linker histone, and the ionic strength. Our results indicate that the linker histone has negligible influence on short NRL fibers, whereas for longer NRL fibers it works like, and in tandem with, concentrated positive counterions to condense the chromatin fiber. Longer NRL fibers also exhibit structural heterogeneity, with solenoid-like conformations viable in addition to irregular zigzags. These features of chromatin and associated internucleosomal patterns presented here help interpret structural dependencies of the chromatin fiber on internal and external factors. In particular, we suggest that longer-NRL are more advantageous for packing and achieving various levels of fiber compaction throughout the cell cycle.
Figures



Similar articles
-
Modeling studies of chromatin fiber structure as a function of DNA linker length.J Mol Biol. 2010 Nov 12;403(5):777-802. doi: 10.1016/j.jmb.2010.07.057. Epub 2010 Aug 13. J Mol Biol. 2010. PMID: 20709077 Free PMC article.
-
Chromatin fiber polymorphism triggered by variations of DNA linker lengths.Proc Natl Acad Sci U S A. 2014 Jun 3;111(22):8061-6. doi: 10.1073/pnas.1315872111. Epub 2014 May 20. Proc Natl Acad Sci U S A. 2014. PMID: 24847063 Free PMC article.
-
Correlation among DNA Linker Length, Linker Histone Concentration, and Histone Tails in Chromatin.Biophys J. 2016 Jun 7;110(11):2309-2319. doi: 10.1016/j.bpj.2016.04.024. Biophys J. 2016. PMID: 27276249 Free PMC article.
-
Chromatosome Structure and Dynamics from Molecular Simulations.Annu Rev Phys Chem. 2020 Apr 20;71:101-119. doi: 10.1146/annurev-physchem-071119-040043. Epub 2020 Feb 4. Annu Rev Phys Chem. 2020. PMID: 32017651 Review.
-
Breaths, Twists, and Turns of Atomistic Nucleosomes.J Mol Biol. 2021 Mar 19;433(6):166744. doi: 10.1016/j.jmb.2020.166744. Epub 2020 Dec 10. J Mol Biol. 2021. PMID: 33309853 Review.
Cited by
-
Brownian dynamics simulations of mesoscale chromatin fibers.Biophys J. 2023 Jul 25;122(14):2884-2897. doi: 10.1016/j.bpj.2022.09.013. Epub 2022 Sep 17. Biophys J. 2023. PMID: 36116007 Free PMC article.
-
Chromatin Compaction Multiscale Modeling: A Complex Synergy Between Theory, Simulation, and Experiment.Front Mol Biosci. 2020 Feb 25;7:15. doi: 10.3389/fmolb.2020.00015. eCollection 2020. Front Mol Biosci. 2020. PMID: 32158765 Free PMC article. Review.
-
Unwinding and rewinding the nucleosome inner turn: force dependence of the kinetic rate constants.Phys Rev E Stat Nonlin Soft Matter Phys. 2013 Jan;87(1):012710. doi: 10.1103/PhysRevE.87.012710. Epub 2013 Jan 17. Phys Rev E Stat Nonlin Soft Matter Phys. 2013. PMID: 23410362 Free PMC article.
-
Nucleosome distribution and linker DNA: connecting nuclear function to dynamic chromatin structure.Biochem Cell Biol. 2011 Feb;89(1):24-34. doi: 10.1139/O10-139. Biochem Cell Biol. 2011. PMID: 21326360 Free PMC article. Review.
-
Structure of native chromatin fibres revealed by Cryo-ET in situ.Nat Commun. 2023 Oct 10;14(1):6324. doi: 10.1038/s41467-023-42072-1. Nat Commun. 2023. PMID: 37816746 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

