Mesoscale simulations of two nucleosome-repeat length oligonucleosomes
- PMID: 20145817
- PMCID: PMC3182521
- DOI: 10.1039/b918629h
Mesoscale simulations of two nucleosome-repeat length oligonucleosomes
Abstract
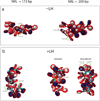
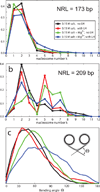
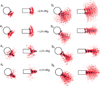
The compaction of chromatin, accessed through coarse-grained modeling and simulation, reveals different folding patterns as a function of the nucleosome repeat length (NRL), the presence of the linker histone, and the ionic strength. Our results indicate that the linker histone has negligible influence on short NRL fibers, whereas for longer NRL fibers it works like, and in tandem with, concentrated positive counterions to condense the chromatin fiber. Longer NRL fibers also exhibit structural heterogeneity, with solenoid-like conformations viable in addition to irregular zigzags. These features of chromatin and associated internucleosomal patterns presented here help interpret structural dependencies of the chromatin fiber on internal and external factors. In particular, we suggest that longer-NRL are more advantageous for packing and achieving various levels of fiber compaction throughout the cell cycle.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

