Dynamic expression of a LEF-EGFP Wnt reporter in mouse development and cancer
- PMID: 20146356
- PMCID: PMC3399184
- DOI: 10.1002/dvg.20604
Dynamic expression of a LEF-EGFP Wnt reporter in mouse development and cancer
Abstract
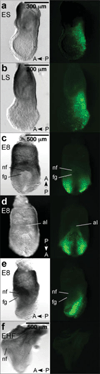
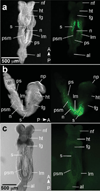
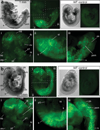
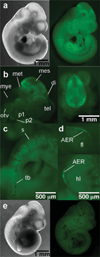
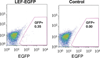
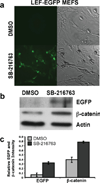
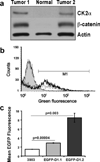
We have characterized a transgenic mouse line in which enhanced green fluorescent protein (EGFP) is expressed under the control of multimerized LEF-1 responsive elements. In embryos, EGFP was detected in known sites of Wnt activation, including the primitive streak, mesoderm, neural tube, somites, heart, limb buds, mammary placodes, and whisker follicles. In vitro cultured transgenic embryonic fibroblasts upregulated EGFP expression in response to activation of Wnt signaling by GSK3beta inhibition. Mammary tumor cell lines derived from female LEF-EGFP transgenic mice treated with the carcinogen 7, 12-dimethylbenz[a]anthracene (DMBA) also express EGFP. Thus, this transgenic line is useful for ex vivo and in vitro studies of Wnt signaling in development and cancer.
(c) 2010 Wiley-Liss, Inc.
Figures


Similar articles
-
Oncogenic signaling pathways activated in DMBA-induced mouse mammary tumors.Toxicol Pathol. 2005;33(6):726-37. doi: 10.1080/01926230500352226. Toxicol Pathol. 2005. PMID: 16263698
-
Activation of different Wnt/beta-catenin signaling components in mammary epithelium induces transdifferentiation and the formation of pilar tumors.Oncogene. 2002 Aug 15;21(36):5548-56. doi: 10.1038/sj.onc.1205686. Oncogene. 2002. PMID: 12165853
-
Regulation of β-catenin nuclear dynamics by GSK-3β involves a LEF-1 positive feedback loop.Traffic. 2011 Aug;12(8):983-99. doi: 10.1111/j.1600-0854.2011.01207.x. Epub 2011 May 13. Traffic. 2011. PMID: 21496192
-
Multifaceted interaction between the androgen and Wnt signaling pathways and the implication for prostate cancer.J Cell Biochem. 2006 Oct 1;99(2):402-10. doi: 10.1002/jcb.20983. J Cell Biochem. 2006. PMID: 16741972 Review.
-
A Wnt-ow of opportunity: targeting the Wnt/beta-catenin pathway in breast cancer.Curr Drug Targets. 2010 Sep;11(9):1074-88. doi: 10.2174/138945010792006780. Curr Drug Targets. 2010. PMID: 20545611 Review.
Cited by
-
Down-regulation of CK2α correlates with decreased expression levels of DNA replication minichromosome maintenance protein complex (MCM) genes.Sci Rep. 2019 Oct 10;9(1):14581. doi: 10.1038/s41598-019-51056-5. Sci Rep. 2019. PMID: 31601942 Free PMC article.
-
Cell Signaling Pathway Reporters in Adult Hematopoietic Stem Cells.Cells. 2020 Oct 9;9(10):2264. doi: 10.3390/cells9102264. Cells. 2020. PMID: 33050292 Free PMC article. Review.
-
Transgenic flash mice for in vivo quantitative monitoring of canonical Wnt signaling to track hair follicle cycle dynamics.J Invest Dermatol. 2014 Jun;134(6):1519-1526. doi: 10.1038/jid.2014.92. Epub 2014 Feb 14. J Invest Dermatol. 2014. PMID: 24531689
-
Myc cooperates with β-catenin to drive gene expression in nephron progenitor cells.Development. 2017 Nov 15;144(22):4173-4182. doi: 10.1242/dev.153700. Epub 2017 Oct 9. Development. 2017. PMID: 28993399 Free PMC article.
-
Sall4 regulates downstream patterning genes during limb regeneration.Dev Biol. 2024 Nov;515:151-159. doi: 10.1016/j.ydbio.2024.07.015. Epub 2024 Jul 25. Dev Biol. 2024. PMID: 39067503
References
-
- Arnold SJ, Stappert J, Bauer A, Kispert A, Herrmann BG, Kemler R. Brachyury is a target gene of the Wnt/beta-catenin signaling pathway. Mech Dev. 2000;91:249–258. - PubMed
-
- Barker N. The canonical Wnt/beta-catenin signalling pathway. Methods Mol Biol. 2008;468:5–15. - PubMed
-
- Barolo S. Transgenic Wnt/TCF pathway reporters: All you need is Lef? Oncogene. 2006;25:7505–7511. - PubMed
-
- Bastidas F, De Calisto J, Mayor R. Identification of neural crest competence territory: Role of Wnt signaling. Dev Dyn. 2004;229:109–117. - PubMed
-
- Boras-Granic K, Chang H, Grosschedl R, Hamel PA. Lef1 is required for the transition of Wnt signaling from mesenchymal to epithelial cells in the mouse embryonic mammary gland. Dev Biol. 2006;295:219–231. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

