Structural characterization of phospholipids and peptides directly from tissue sections by MALDI traveling-wave ion mobility-mass spectrometry
- PMID: 20146447
- PMCID: PMC2830338
- DOI: 10.1021/ac9026115
Structural characterization of phospholipids and peptides directly from tissue sections by MALDI traveling-wave ion mobility-mass spectrometry
Abstract
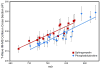
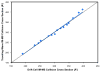
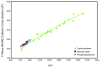
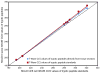
Ion mobility-mass spectrometry (IM-MS) provides rapid two-dimensional separations based on analyte apparent surface area or collision cross section (CCS, A(2)) and mass-to-charge, respectively. Recently, traveling-wave (t-wave) IM-MS was developed which uses electrodynamic rather than electrostatic fields commonly used in drift cell IM-MS instruments. The underlying theory for obtaining CCS data is well developed for drift cell IM-MS, while strategies for obtaining CCS values from t-wave IM-MS data remains an active area of research. In this report, methods were developed and validated to obtain CCS values of phospholipids and peptides directly from thin tissue sections by MALDI t-wave IM-MS using CCS calibrants measured by MALDI drift cell IM-MS. Importantly, the average percent difference between t-wave and drift cell CCS measurements is minimized by calibrating with the same biomolecular class. Calibrating t-wave phospholipid CCS values with drift cell peptide CCS measurements results in an average percent difference of ca. 7% between the same lipids measured using t-wave and drift cell IM-MS, while this improves to <0.5% when drift cell phospholipid CCS values are used for calibrating t-wave data. A suite of CCS values are reported for lipids and peptides that were determined directly from tissue, i.e. without the need for tissue extraction and further purification steps.
Figures



References
-
- McLean JA, Ruotolo BT, Gillig KJ, Russell DH. International Journal of Mass Spectrometry. 2005;240:301–315.
-
- Valentine SJ, Liu XY, Plasencia MD, Hilderbrand AE, Kurulugama RT, Koeniger SL, Clemmer DE. Expert Review of Proteomics. 2005;2:553–565. - PubMed
-
- Wyttenbach T, Bowers MT. Annual Review of Physical Chemistry. 2007;58:511–533. - PubMed
-
- Kanu AB, Dwivedi P, Tam M, Matz L, Hill HH., Jr Journal of Mass Spectrometry. 2008;43:1–22. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

