Ion selectivity of alpha-hemolysin with beta-cyclodextrin adapter. II. Multi-ion effects studied with grand canonical Monte Carlo/Brownian dynamics simulations
- PMID: 20146515
- PMCID: PMC2843906
- DOI: 10.1021/jp906791b
Ion selectivity of alpha-hemolysin with beta-cyclodextrin adapter. II. Multi-ion effects studied with grand canonical Monte Carlo/Brownian dynamics simulations
Abstract
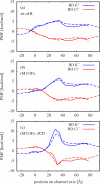
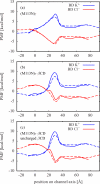
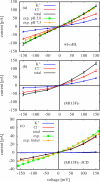
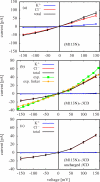
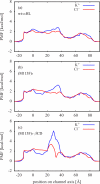
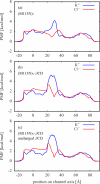
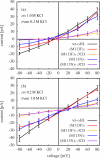
In a previous study of ion selectivity of alpha-hemolysin (alphaHL) in complex with beta-cyclodextrin (betaCD) adapter, we calculated the potential of mean force (PMF) and characterized the self-diffusion coefficients of isolated K(+) and Cl(-) ions using molecular dynamics simulations (Y. Luo et al., "Ion Selectivity of alpha-Hemolysin with beta-Cyclodextrin Adapter: I. Single Ion Potential of Mean Force and Diffusion Coefficient"). In the present effort, these results pertaining to single isolated ions in the wide aqueous pore are extended to take into account multi-ion effects. The grand canonical Monte Carlo/Brownian dynamics (GCMC/BD) algorithm is used to simulate ion currents through the wild-type alphaHL ion channel, as well as two engineered alphaHL mutants, with and without the cyclic oligosaccaride betaCD lodged in the lumen of the pore. The GCMC/BD current-voltage curves agree well with experimental results and show that betaCD increases the anion selectivity of alphaHL. Comparisons between multi-ion PMFs from GCMC/BD simulations and single-ion PMFs demonstrate that multi-ion effects and pore shape are crucial for explaining this behavior. It is concluded that the narrow betaCD adapter increases the anion selectivity of alphaHL because it reduces the pore radius locally, which decreases the ionic screening and the dielectric shielding of the strong electrostatic field induced by a nearby ring of positively charged alphaHL side chains.
Figures

Similar articles
-
Ion selectivity of alpha-hemolysin with a beta-cyclodextrin adapter. I. Single ion potential of mean force and diffusion coefficient.J Phys Chem B. 2010 Jan 21;114(2):952-8. doi: 10.1021/jp906790f. J Phys Chem B. 2010. PMID: 20041673 Free PMC article.
-
Reversal of charge selectivity in transmembrane protein pores by using noncovalent molecular adapters.Proc Natl Acad Sci U S A. 2000 Apr 11;97(8):3959-64. doi: 10.1073/pnas.97.8.3959. Proc Natl Acad Sci U S A. 2000. PMID: 10760267 Free PMC article.
-
Interaction of the noncovalent molecular adapter, beta-cyclodextrin, with the staphylococcal alpha-hemolysin pore.Biophys J. 2000 Oct;79(4):1967-75. doi: 10.1016/S0006-3495(00)76445-9. Biophys J. 2000. PMID: 11023901 Free PMC article.
-
Prolonged residence time of a noncovalent molecular adapter, beta-cyclodextrin, within the lumen of mutant alpha-hemolysin pores.J Gen Physiol. 2001 Nov;118(5):481-94. doi: 10.1085/jgp.118.5.481. J Gen Physiol. 2001. PMID: 11696607 Free PMC article.
-
Beta-barrel membrane protein folding and structure viewed through the lens of alpha-hemolysin.Biochim Biophys Acta. 2003 Jan 10;1609(1):19-27. doi: 10.1016/s0005-2736(02)00663-6. Biochim Biophys Acta. 2003. PMID: 12507754 Review.
Cited by
-
Membrane Position Dependency of the pKa and Conductivity of the Protein Ion Channel.J Membr Biol. 2018 Jun;251(3):393-404. doi: 10.1007/s00232-018-0013-3. Epub 2018 Jan 16. J Membr Biol. 2018. PMID: 29340712 Free PMC article.
-
Molecular dynamics studies of ion permeation in VDAC.Biophys J. 2011 Feb 2;100(3):602-610. doi: 10.1016/j.bpj.2010.12.3711. Biophys J. 2011. PMID: 21281574 Free PMC article.
-
Cyclodextrins: Only Pharmaceutical Excipients or Full-Fledged Drug Candidates?Pharmaceutics. 2022 Nov 22;14(12):2559. doi: 10.3390/pharmaceutics14122559. Pharmaceutics. 2022. PMID: 36559052 Free PMC article. Review.
-
Computational investigation of cholesterol binding sites on mitochondrial VDAC.J Phys Chem B. 2014 Aug 21;118(33):9852-60. doi: 10.1021/jp504516a. Epub 2014 Aug 11. J Phys Chem B. 2014. PMID: 25080204 Free PMC article.
-
Molecular bases of cyclodextrin adapter interactions with engineered protein nanopores.Proc Natl Acad Sci U S A. 2010 May 4;107(18):8165-70. doi: 10.1073/pnas.0914229107. Epub 2010 Apr 16. Proc Natl Acad Sci U S A. 2010. PMID: 20400691 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

