Antigen identification and characterization of lung cancer specific monoclonal antibodies produced by mAb proteomics
- PMID: 20146545
- PMCID: PMC2849899
- DOI: 10.1021/pr900997z
Antigen identification and characterization of lung cancer specific monoclonal antibodies produced by mAb proteomics
Abstract
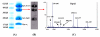
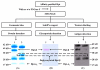
A mass spectrometric (MS)-based strategy for antigen (Ag) identification and characterization of globally produced monoclonal antibodies (mAbs) is described. Mice were immunized with a mixture of native glycoproteins, isolated from the pooled plasma of patients with nonsmall cell lung cancer (NSCLC), to generate a library of IgG-secreting hybridomas. Prior to immunization, the pooled NSCLC plasma was subjected to 3-sequential steps of affinity fractionation, including high abundant plasma protein depletion, glycoprotein enrichment, and polyclonal antibody affinity chromatography normalization. In this paper, to demonstrate the high quality of the globally produced mAbs, we selected 3 mAbs of high differentiating power against a matched, pooled normal plasma sample. After production of large quantities of the mAbs from ascites fluids, Ag identification was achieved by immunoaffinity purification, SDS-PAGE, Western blotting, and MS analysis of in-gel digest products. One antigen was found to be complement factor H, and the other two were mapped to different subunits of haptoglobin (Hpt). The 2 Hpt mAbs were characterized in detail to assess the quality of the mAbs produced by the global strategy. The affinity of one of the mAbs to the Hpt native tetramer form was found to have a K(D) of roughly 10(-9) M and to be 2 orders of magnitude lower than the reduced form, demonstrating the power of the mAb proteomics technology in generating mAbs to the natural form of the proteins in blood. The binding of this mAb to the beta-chain of haptoglobin was also dependent on glycosylation on this chain. The characterization of mAbs in this work reveals that the global mAb proteomics process can generate high-quality lung cancer specific mAbs capable of recognizing proteins in their native state.
Figures

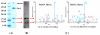


Similar articles
-
Discovery of lung cancer biomarkers by profiling the plasma proteome with monoclonal antibody libraries.Mol Cell Proteomics. 2011 Dec;10(12):M111.010298. doi: 10.1074/mcp.M111.010298. Epub 2011 Sep 26. Mol Cell Proteomics. 2011. PMID: 21947365 Free PMC article.
-
Delineating monoclonal antibody specificity by mass spectrometry.J Proteomics. 2015 Jan 30;114:115-24. doi: 10.1016/j.jprot.2014.11.004. Epub 2014 Nov 15. J Proteomics. 2015. PMID: 25462431
-
An alternative strategy for high throughput generation and characterization of monoclonal antibodies against human plasma proteins using fractionated native proteins as immunogens.Proteomics. 2006 Jan;6(2):438-48. doi: 10.1002/pmic.200500327. Proteomics. 2006. PMID: 16419015
-
Ultrasensitive characterization of site-specific glycosylation of affinity-purified haptoglobin from lung cancer patient plasma using 10 μm i.d. porous layer open tubular liquid chromatography-linear ion trap collision-induced dissociation/electron transfer dissociation mass spectrometry.Anal Chem. 2011 Mar 15;83(6):2029-37. doi: 10.1021/ac102825g. Epub 2011 Feb 21. Anal Chem. 2011. PMID: 21338062 Free PMC article.
-
Rat plasma proteomics: effects of abundant protein depletion on proteomic analysis.J Chromatogr B Analyt Technol Biomed Life Sci. 2007 Apr 15;849(1-2):273-81. doi: 10.1016/j.jchromb.2006.11.051. Epub 2006 Dec 22. J Chromatogr B Analyt Technol Biomed Life Sci. 2007. PMID: 17188586
Cited by
-
The study on newly developed McAb NJ001 specific to non-small cell lung cancer and its biological characteristics.PLoS One. 2012;7(3):e33009. doi: 10.1371/journal.pone.0033009. Epub 2012 Mar 30. PLoS One. 2012. PMID: 22479355 Free PMC article.
-
Large-Scale Plasma Proteome Epitome Profiling is an Efficient Tool for the Discovery of Cancer Biomarkers.Mol Cell Proteomics. 2023 Jul;22(7):100580. doi: 10.1016/j.mcpro.2023.100580. Epub 2023 May 20. Mol Cell Proteomics. 2023. PMID: 37211046 Free PMC article.
-
Dissecting virus-plant interactions through proteomics approaches.Curr Proteomics. 2010 Dec 1;7(4):316-327. doi: 10.2174/157016410793611792. Curr Proteomics. 2010. PMID: 24039615 Free PMC article.
-
Application of an antibody chip for screening differentially expressed proteins during peach ripening and identification of a metabolon in the SAM cycle to generate a peach ethylene biosynthesis model.Hortic Res. 2020 Mar 15;7:31. doi: 10.1038/s41438-020-0249-9. eCollection 2020. Hortic Res. 2020. PMID: 32194967 Free PMC article.
-
Discovery of lung cancer biomarkers by profiling the plasma proteome with monoclonal antibody libraries.Mol Cell Proteomics. 2011 Dec;10(12):M111.010298. doi: 10.1074/mcp.M111.010298. Epub 2011 Sep 26. Mol Cell Proteomics. 2011. PMID: 21947365 Free PMC article.
References
-
- Werner ZJ, Hanno L. How industry is approaching the search for new diagnostic markers and biomarkers. Mol. Cell. Proteomics. 2004;3:345–354. - PubMed
-
- Chen R, Pan S, Brentnall TA, Aebersold R. Proteomic profiling of pancreatic cancer for biomarker discovery. Mol. Cell. Proteomics. 2005;4:523–533. - PubMed
-
- Rifai N, Gillette MA, Carr SA. Protein biomarker discovery and validation: the long and uncertain path to clinical utility. Nat. Biotech. 2006;24:971–983. - PubMed
-
- Goldsby R, Kindt T, Osborne B. Kuby immunology. 4th edition. W.H. Freeman publishing; 2002. pp. 148–150. chapter 6.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

