Identification and analysis of in planta expressed genes of Magnaporthe oryzae
- PMID: 20146797
- PMCID: PMC2832786
- DOI: 10.1186/1471-2164-11-104
Identification and analysis of in planta expressed genes of Magnaporthe oryzae
Abstract
Background: Infection of plants by pathogens and the subsequent disease development involves substantial changes in the biochemistry and physiology of both partners. Analysis of genes that are expressed during these interactions represents a powerful strategy to obtain insights into the molecular events underlying these changes. We have employed expressed sequence tag (EST) analysis to identify rice genes involved in defense responses against infection by the blast fungus Magnaporthe oryzae and fungal genes involved in infectious growth within the host during a compatible interaction.
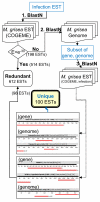
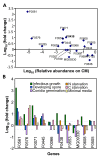
Results: A cDNA library was constructed with RNA from rice leaves (Oryza sativa cv. Hwacheong) infected with M. oryzae strain KJ201. To enrich for fungal genes, subtraction library using PCR-based suppression subtractive hybridization was constructed with RNA from infected rice leaves as a tester and that from uninfected rice leaves as the driver. A total of 4,148 clones from two libraries were sequenced to generate 2,302 non-redundant ESTs. Of these, 712 and 1,562 ESTs could be identified to encode fungal and rice genes, respectively. To predict gene function, Gene Ontology (GO) analysis was applied, with 31% and 32% of rice and fungal ESTs being assigned to GO terms, respectively. One hundred uniESTs were found to be specific to fungal infection EST. More than 80 full-length fungal cDNA sequences were used to validate ab initio annotated gene model of M. oryzae genome sequence.
Conclusion: This study shows the power of ESTs to refine genome annotation and functional characterization. Results of this work have advanced our understanding of the molecular mechanisms underpinning fungal-plant interactions and formed the basis for new hypothesis.
Figures




Similar articles
-
Analysis of genes expressed during rice-Magnaporthe grisea interactions.Mol Plant Microbe Interact. 2001 Nov;14(11):1340-6. doi: 10.1094/MPMI.2001.14.11.1340. Mol Plant Microbe Interact. 2001. PMID: 11763134
-
Large-scale identification of expressed sequence tags involved in rice and rice blast fungus interaction.Plant Physiol. 2005 May;138(1):105-15. doi: 10.1104/pp.104.055624. Plant Physiol. 2005. PMID: 15888683 Free PMC article.
-
Identification of fungal ( Magnaporthe grisea) stress-induced genes in wild rice ( Oryza minuta).Plant Cell Rep. 2004 Mar;22(8):599-607. doi: 10.1007/s00299-003-0742-2. Epub 2003 Nov 27. Plant Cell Rep. 2004. PMID: 14648106
-
Gene Ontology annotation of the rice blast fungus, Magnaporthe oryzae.BMC Microbiol. 2009 Feb 19;9 Suppl 1(Suppl 1):S8. doi: 10.1186/1471-2180-9-S1-S8. BMC Microbiol. 2009. PMID: 19278556 Free PMC article. Review.
-
Recent progress and understanding of the molecular mechanisms of the rice-Magnaporthe oryzae interaction.Mol Plant Pathol. 2010 May;11(3):419-27. doi: 10.1111/j.1364-3703.2009.00607.x. Mol Plant Pathol. 2010. PMID: 20447289 Free PMC article. Review.
Cited by
-
Genome plasticity in filamentous plant pathogens contributes to the emergence of novel effectors and their cellular processes in the host.Curr Genet. 2016 Feb;62(1):47-51. doi: 10.1007/s00294-015-0509-7. Epub 2015 Jul 31. Curr Genet. 2016. PMID: 26228744 Review.
-
Comparative analysis of zinc finger proteins involved in plant disease resistance.PLoS One. 2012;7(8):e42578. doi: 10.1371/journal.pone.0042578. Epub 2012 Aug 15. PLoS One. 2012. PMID: 22916136 Free PMC article.
-
Comparative genome analyses reveal sequence features reflecting distinct modes of host-adaptation between dicot and monocot powdery mildew.BMC Genomics. 2018 Sep 25;19(1):705. doi: 10.1186/s12864-018-5069-z. BMC Genomics. 2018. PMID: 30253736 Free PMC article.
-
Global expression profiling of transcription factor genes provides new insights into pathogenicity and stress responses in the rice blast fungus.PLoS Pathog. 2013;9(6):e1003350. doi: 10.1371/journal.ppat.1003350. Epub 2013 Jun 6. PLoS Pathog. 2013. PMID: 23762023 Free PMC article.
-
Regulation of cellular diacylglycerol through lipid phosphate phosphatases is required for pathogenesis of the rice blast fungus, Magnaporthe oryzae.PLoS One. 2014 Jun 24;9(6):e100726. doi: 10.1371/journal.pone.0100726. eCollection 2014. PLoS One. 2014. PMID: 24959955 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

