Genome-wide analysis of the diatom cell cycle unveils a novel type of cyclins involved in environmental signaling
- PMID: 20146805
- PMCID: PMC2872877
- DOI: 10.1186/gb-2010-11-2-r17
Genome-wide analysis of the diatom cell cycle unveils a novel type of cyclins involved in environmental signaling
Abstract
Background: Despite the enormous importance of diatoms in aquatic ecosystems and their broad industrial potential, little is known about their life cycle control. Diatoms typically inhabit rapidly changing and unstable environments, suggesting that cell cycle regulation in diatoms must have evolved to adequately integrate various environmental signals. The recent genome sequencing of Thalassiosira pseudonana and Phaeodactylum tricornutum allows us to explore the molecular conservation of cell cycle regulation in diatoms.
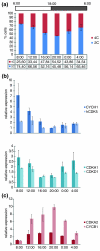
Results: By profile-based annotation of cell cycle genes, counterparts of conserved as well as new regulators were identified in T. pseudonana and P. tricornutum. In particular, the cyclin gene family was found to be expanded extensively compared to that of other eukaryotes and a novel type of cyclins was discovered, the diatom-specific cyclins. We established a synchronization method for P. tricornutum that enabled assignment of the different annotated genes to specific cell cycle phase transitions. The diatom-specific cyclins are predominantly expressed at the G1-to-S transition and some respond to phosphate availability, hinting at a role in connecting cell division to environmental stimuli.
Conclusion: The discovery of highly conserved and new cell cycle regulators suggests the evolution of unique control mechanisms for diatom cell division, probably contributing to their ability to adapt and survive under highly fluctuating environmental conditions.
Figures



References
-
- Hoek C Van den, Mann DG, Jahns HM. Algae: An Introduction to Phycology. Cambridge: Cambridge University Press; 1995.
-
- Kooistra WH, De Stefano M, Mann DG, Medlin LK. The phylogeny of the diatoms. Prog Mol Subcell Biol. 2003;33:59–97. - PubMed
-
- Armbrust EV, Berges JA, Bowler C, Green BR, Martinez D, Putnam NH, Zhou SG, Allen AE, Apt KE, Bechner M, Brzezinski MA, Chaal BK, Chiovitti A, Davis AK, Demarest MS, Detter JC, Glavina T, Goodstein D, Hadi MZ, Hellsten U, Hildebrand M, Jenkins BD, Jurka J, Kapitonov VV, Kröger N, Lau WWY, Lane TW, Larimer FW, Lippmeier JC, Lucas S. The genome of the diatom Thalassiosira pseudonana: Ecology, evolution, and metabolism. Science. 2004;306:79–86. doi: 10.1126/science.1101156. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

