Role of apoptosis-inducing factor (AIF) in programmed nuclear death during conjugation in Tetrahymena thermophila
- PMID: 20146827
- PMCID: PMC2829475
- DOI: 10.1186/1471-2121-11-13
Role of apoptosis-inducing factor (AIF) in programmed nuclear death during conjugation in Tetrahymena thermophila
Abstract
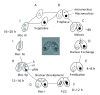
Background: Programmed nuclear death (PND), which is also referred to as nuclear apoptosis, is a remarkable process that occurs in ciliates during sexual reproduction (conjugation). In Tetrahymena thermophila, when the new macronucleus differentiates, the parental macronucleus is selectively eliminated from the cytoplasm of the progeny, concomitant with apoptotic nuclear events. However, the molecular mechanisms underlying these events are not well understood. The parental macronucleus is engulfed by a large autophagosome, which contains numerous mitochondria that have lost their membrane potential. In animals, mitochondrial depolarization precedes apoptotic cell death, which involves DNA fragmentation and subsequent nuclear degradation.
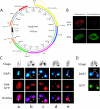
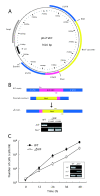
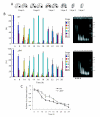
Results: We focused on the role of mitochondrial apoptosis-inducing factor (AIF) during PND in Tetrahymena. The disruption of AIF delays the normal progression of PND, specifically, nuclear condensation and kilobase-size DNA fragmentation. AIF is localized in Tetrahymena mitochondria and is released into the macronucleus prior to nuclear condensation. In addition, AIF associates and co-operates with the mitochondrial DNase to facilitate the degradation of kilobase-size DNA, which is followed by oligonucleosome-size DNA laddering.
Conclusions: Our results suggest that Tetrahymena AIF plays an important role in the degradation of DNA at an early stage of PND, which supports the notion that the mitochondrion-initiated apoptotic DNA degradation pathway is widely conserved among eukaryotes.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

