SoDA2: a Hidden Markov Model approach for identification of immunoglobulin rearrangements
- PMID: 20147303
- PMCID: PMC2844993
- DOI: 10.1093/bioinformatics/btq056
SoDA2: a Hidden Markov Model approach for identification of immunoglobulin rearrangements
Abstract
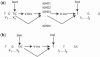
Motivation: The inference of pre-mutation immunoglobulin (Ig) rearrangements is essential in the study of the antibody repertoires produced in response to infection, in B-cell neoplasms and in autoimmune disease. Often, there are several rearrangements that are nearly equivalent as candidates for a given Ig gene, but have different consequences in an analysis. Our aim in this article is to develop a probabilistic model of the rearrangement process and a Bayesian method for estimating posterior probabilities for the comparison of multiple plausible rearrangements.
Results: We have developed SoDA2, which is based on a Hidden Markov Model and used to compute the posterior probabilities of candidate rearrangements and to find those with the highest values among them. We validated the software on a set of simulated data, a set of clonally related sequences, and a group of randomly selected Ig heavy chains from Genbank. In most tests, SoDA2 performed better than other available software for the task. Furthermore, the output format has been redesigned, in part, to facilitate comparison of multiple solutions.
Availability: SoDA2 is available online at https://hippocrates.duhs.duke.edu/soda. Simulated sequences are available upon request.
Figures


References
-
- Altschul SF, et al. Basic local alignment tool. J. Mol. Biol. 1990;215:403–410. - PubMed
-
- Basu M, et al. Synthesis of compositionally unique DNA by terminal deoxynucleotidyl transferase. Biochem. Biophys. Res. Commun. 1983;111:1105–1112. - PubMed
-
- Cowell LG, et al. Enhanced evolvability in immunoglobulin V genes under somatic hypermutation. J. Mol. Evol. 1999;49:23–26. - PubMed
-
- Desiderio SV, et al. Insertion of N regions into heavy-chain genes is correlated with the expression of terminal deoxytransferase in B-cells. Nature. 1984;311:752–757. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

