Protocols for sampling viral sequences to study epidemic dynamics
- PMID: 20147314
- PMCID: PMC2880085
- DOI: 10.1098/rsif.2009.0530
Protocols for sampling viral sequences to study epidemic dynamics
Abstract
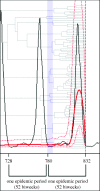
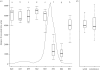
With more emphasis being put on global infectious disease monitoring, viral genetic data are being collected at an astounding rate, both within and without the context of a long-term disease surveillance plan. Concurrent with this increase have come improvements to the sophisticated and generalized statistical techniques used for extracting population-level information from genetic sequence data. However, little research has been done on how the collection of these viral sequence data can or does affect the efficacy of the phylogenetic algorithms used to analyse and interpret them. In this study, we use epidemic simulations to consider how the collection of viral sequence data clarifies or distorts the picture, provided by the phylogenetic algorithms, of the underlying population dynamics of the simulated viral infection over many epidemic cycles. We find that sampling protocols purposefully designed to capture sequences at specific points in the epidemic cycle, such as is done for seasonal influenza surveillance, lead to a significantly better view of the underlying population dynamics than do less-focused collection protocols. Our results suggest that the temporal distribution of samples can have a significant effect on what can be inferred from genetic data, and thus highlight the importance of considering this distribution when designing or evaluating protocols and analysing the data collected thereunder.
Figures


References
-
- Bjornstad O. N., Finkenstadt B. F., Grenfell B. T. 2002. Dynamics of measles epidemics: estimating scaling of transmission rates using a time series SIR model. Ecol. Monogr. 72, 169–184.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
