Structure-based mutational analysis of the bovine papillomavirus E1 helicase domain identifies residues involved in the nonspecific DNA binding activity required for double trimer formation
- PMID: 20147403
- PMCID: PMC2863750
- DOI: 10.1128/JVI.02214-09
Structure-based mutational analysis of the bovine papillomavirus E1 helicase domain identifies residues involved in the nonspecific DNA binding activity required for double trimer formation
Abstract
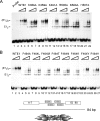
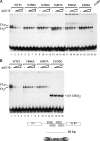
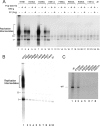
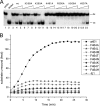
The papillomavirus E1 protein is a multifunctional initiator protein responsible for preparing the viral DNA template for initiation of DNA replication. The E1 protein encodes two DNA binding activities that are required for initiation of DNA replication. A well-characterized sequence-specific DNA binding activity resides in the E1 DBD and is used to tether E1 to the papillomavirus ori. A non-sequence-specific DNA binding activity is also required for formation of the E1 double trimer (DT) complex, which is responsible for the local template melting that precedes loading of the E1 helicase. This DNA binding activity is very poorly understood. We use a structure-based mutagenesis approach to identify residues in the E1 helicase domain that are required for the non-sequence-specific DNA binding and DT formation. We found that three groups of residues are involved in nonspecific DNA binding: the E1 beta-hairpin structure containing R505, K506, and H507; a hydrophobic loop containing F464; and a charged loop containing K461 together generate the binding surface involved in nonspecific DNA binding. These residues are well conserved in the T antigens from the polyomaviruses, indicating that the polyomaviruses share this nonspecific DNA binding activity.
Figures




Similar articles
-
A DNA-binding activity in BPV initiator protein E1 required for melting duplex ori DNA but not processive helicase activity initiated on partially single-stranded DNA.Nucleic Acids Res. 2008 Apr;36(6):1891-9. doi: 10.1093/nar/gkn041. Epub 2008 Feb 11. Nucleic Acids Res. 2008. PMID: 18267969 Free PMC article.
-
Common determinants in DNA melting and helicase-catalysed DNA unwinding by papillomavirus replication protein E1.Nucleic Acids Res. 2006 May 31;34(10):3008-19. doi: 10.1093/nar/gkl384. Print 2006. Nucleic Acids Res. 2006. PMID: 16738139 Free PMC article.
-
DNA-binding domain of bovine papillomavirus type 1 E1 helicase: structural and functional aspects.J Virol. 1993 Oct;67(10):6000-14. doi: 10.1128/JVI.67.10.6000-6014.1993. J Virol. 1993. PMID: 8396665 Free PMC article.
-
Papillomavirus E1 proteins: form, function, and features.Virus Genes. 2002 Jun;24(3):275-90. doi: 10.1023/a:1015336817836. Virus Genes. 2002. PMID: 12086149 Review.
-
Genetic Variability of the Functional Domains of Chromodomains Helicase DNA-Binding (CHD) Proteins.Genes (Basel). 2021 Nov 19;12(11):1827. doi: 10.3390/genes12111827. Genes (Basel). 2021. PMID: 34828433 Free PMC article. Review.
Cited by
-
Nuclear export of human papillomavirus type 31 E1 is regulated by Cdk2 phosphorylation and required for viral genome maintenance.J Virol. 2010 Nov;84(22):11747-60. doi: 10.1128/JVI.01445-10. Epub 2010 Sep 15. J Virol. 2010. PMID: 20844047 Free PMC article.
-
The inhibitory action of P56 on select functions of E1 mediates interferon's effect on human papillomavirus DNA replication.J Virol. 2010 Dec;84(24):13036-9. doi: 10.1128/JVI.01194-10. Epub 2010 Oct 6. J Virol. 2010. PMID: 20926571 Free PMC article.
-
Complete genome sequences of three novel human papillomavirus types, 175, 178, and 180.Genome Announc. 2014 May 22;2(3):e00443-14. doi: 10.1128/genomeA.00443-14. Genome Announc. 2014. PMID: 24855297 Free PMC article.
-
In planta production of a candidate vaccine against bovine papillomavirus type 1.Planta. 2012 Oct;236(4):1305-13. doi: 10.1007/s00425-012-1692-0. Epub 2012 Jun 21. Planta. 2012. PMID: 22718313
-
A conserved regulatory module at the C terminus of the papillomavirus E1 helicase domain controls E1 helicase assembly.J Virol. 2015 Jan 15;89(2):1129-42. doi: 10.1128/JVI.01903-14. Epub 2014 Nov 5. J Virol. 2015. PMID: 25378487 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

