Internal and external paralogy in the evolution of tropomyosin genes in metazoans
- PMID: 20147436
- PMCID: PMC2912468
- DOI: 10.1093/molbev/msq018
Internal and external paralogy in the evolution of tropomyosin genes in metazoans
Abstract
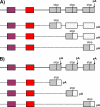
Nature contains a tremendous diversity of forms both at the organismal and genomic levels. This diversity motivates the twin central questions of molecular evolution: what are the molecular mechanisms of adaptation, and what are the functional consequences of genomic diversity. We report a 22-species comparative analysis of tropomyosin (PPM) genes, which exist in a variety of forms and are implicated in the emergence of a wealth of cellular functions, including the novel muscle functions integral to the functional diversification of bilateral animals. TPM genes encode either or both of long-form [284 amino acid (aa)] and short-form (approximately 248 aa) proteins. Consistent with a role of TPM diversification in the origins and radiation of bilaterians, we find evidence that the muscle-specific long-form protein arose in proximal bilaterian ancestors (the bilaterian 'stem'). Duplication of the 5' end of the gene led to alternative promoters encoding long- and short-form transcripts with distinct functions. This dual-function gene then underwent strikingly parallel evolution in different bilaterian lineages. In each case, recurrent tandem exon duplication and mutually exclusive alternative splicing of the duplicates, with further association between these alternatively spliced exons along the gene, led to long- and short-form-specific exons, allowing for gradual emergence of alternative "internal paralogs" within the same gene. We term these Mutually exclusively Alternatively spliced Tandemly duplicated Exon sets "MATEs". This emergence of internal paralogs in various bilaterians has employed every single TPM exon in at least one lineage and reaches striking levels of divergence with up to 77% of long- and short-form transcripts being transcribed from different genomic regions. Interestingly, in some lineages, these internal alternatively spliced paralogs have subsequently been "externalized" by full gene duplication and reciprocal retention/loss of the two transcript isoforms, a particularly clear case of evolution by subfunctionalization. This parallel evolution of TPM genes in diverse metazoans attests to common selective forces driving divergence of different gene transcripts and represents a striking case of emergence of evolutionary novelty by alternative splicing.
Figures




Similar articles
-
Three novel brain tropomyosin isoforms are expressed from the rat alpha-tropomyosin gene through the use of alternative promoters and alternative RNA processing.Mol Cell Biol. 1990 Apr;10(4):1729-42. doi: 10.1128/mcb.10.4.1729-1742.1990. Mol Cell Biol. 1990. PMID: 2320008 Free PMC article.
-
Extensive divergence in alternative splicing patterns after gene and genome duplication during the evolutionary history of Arabidopsis.Mol Biol Evol. 2010 Jul;27(7):1686-97. doi: 10.1093/molbev/msq054. Epub 2010 Feb 25. Mol Biol Evol. 2010. PMID: 20185454
-
Evidence for widespread subfunctionalization of splice forms in vertebrate genomes.Genome Res. 2015 May;25(5):624-32. doi: 10.1101/gr.184473.114. Epub 2015 Mar 19. Genome Res. 2015. PMID: 25792610 Free PMC article.
-
Evolutionary convergence of alternative splicing in ion channels.Trends Genet. 2004 Apr;20(4):171-6. doi: 10.1016/j.tig.2004.02.001. Trends Genet. 2004. PMID: 15101391 Review.
-
Tropomyosin exons as models for alternative splicing.Adv Exp Med Biol. 2008;644:27-42. doi: 10.1007/978-0-387-85766-4_3. Adv Exp Med Biol. 2008. PMID: 19209811 Review.
Cited by
-
ExOrthist: a tool to infer exon orthologies at any evolutionary distance.Genome Biol. 2021 Aug 20;22(1):239. doi: 10.1186/s13059-021-02441-9. Genome Biol. 2021. PMID: 34416914 Free PMC article.
-
Molecular architecture of muscles in an acoel and its evolutionary implications.J Exp Zool B Mol Dev Evol. 2011 Sep 15;316(6):427-39. doi: 10.1002/jez.b.21416. Epub 2011 May 2. J Exp Zool B Mol Dev Evol. 2011. PMID: 21538843 Free PMC article.
-
Origins and Evolution of Human Tandem Duplicated Exon Substitution Events.Genome Biol Evol. 2022 Dec 7;14(12):evac162. doi: 10.1093/gbe/evac162. Genome Biol Evol. 2022. PMID: 36346145 Free PMC article.
-
Predicting the impact of alternative splicing on plant MADS domain protein function.PLoS One. 2012;7(1):e30524. doi: 10.1371/journal.pone.0030524. Epub 2012 Jan 25. PLoS One. 2012. PMID: 22295091 Free PMC article.
-
The role of tropomyosin domains in cooperative activation of the actin-myosin interaction.J Mol Biol. 2011 Dec 16;414(5):667-80. doi: 10.1016/j.jmb.2011.10.026. Epub 2011 Oct 20. J Mol Biol. 2011. PMID: 22041451 Free PMC article.
References
-
- Abascal F, Zardoya R, Posada D. ProtTest: selection of best-fit models of protein evolution. Bioinformatics. 2005;21:2104–2105. - PubMed
-
- Blanchard EM, Iizuka K, Christe M, Conner DA, Geisterfer-Lowrance A, Schoen FJ, Maughan DW, Seidman CE, Seidman JG. Targeted ablation of the murine alpha-tropomyosin gene. Circ Res. 1997;81:1005–1010. - PubMed
-
- Brites D, McTaggart S, Morris K, Anderson J, Thomas K, Colson I, Fabbro T, Little T, Ebert D, Du Pasquier L. The Dscam homologue of the Crustacean Daphnia is diversified by alternative splicing like in insects. Mol Biol Evol. 2008;25:1429–1439. - PubMed
-
- D'Aniello S, Irimia M, Maeso I, Pascual-Anaya J, Jiménez-Delgado S, Bertrand S, Garcia-Fernàndez J. Gene expansion and retention leads to a diverse tyrosine kinase superfamily in amphioxus. Mol Biol Evol. 2008;25:1841–1854. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

