Translocation of Y-linked genes to the dot chromosome in Drosophila pseudoobscura
- PMID: 20147437
- PMCID: PMC2912472
- DOI: 10.1093/molbev/msq045
Translocation of Y-linked genes to the dot chromosome in Drosophila pseudoobscura
Abstract
One of the most striking cases of sex chromosome reorganization in Drosophila occurred in the lineage ancestral to Drosophila pseudoobscura, where there was a translocation of Y-linked genes to an autosome. These genes went from being present only in males, never recombining, and having an effective population size of 0.5N to a state of autosomal linkage, where they are passed through both sexes, may recombine, and their effective population size has quadrupled. These genes appear to be functional, and they underwent a drastic reduction in intron size after the translocation. A Y-autosome translocation may pose problems in meiosis if the rDNA locus responsible for X-Y pairing had also moved to an autosome. In this study, we demonstrate that the Y-autosome translocation moved Y-linked genes onto the dot chromosome, a small, mainly heterochromatic autosome with some sex chromosome-like properties. The rDNA repeats occur exclusively on the X chromosome in D. pseudoobscura, but we found that the new Y chromosome of this species harbors four clusters bearing only the intergenic spacer region (IGS) of the rDNA repeats. This arrangement appears analogous to the situation in Drosophila simulans, where X-rDNA to Y-IGS pairing could be responsible for X-Y chromosome pairing. We postulate that the nascent D. pseudoobscura Y chromosome acquired and amplified copies of the IGS, suggesting a potential mechanism for X-Y pairing in D. pseudoobscura.
Figures
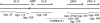
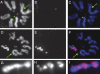
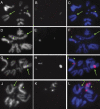



References
-
- Ashburner M. Drosophila. a laboratory handbook. Cold Spring Harbor (NY): Cold Spring Harbor Laboratory Press; 1989.
-
- Ault JG, Rieder CL. Meiosis in Drosophila males. I. The question of separate conjunctive mechanisms for the XY and autosomal bivalents. Chromosoma. 1994;103:352–356. - PubMed
-
- Brianti MT, Ananina G, Recco-Pimentel SM, Klaczko LB. Comparative analysis of the chromosomal positions of rDNA genes in species of the tripunctata radiation of Drosophila. Cytogenet Genome Res. 2009;125:149–157. - PubMed
-
- Bridges CB. The mutants and linkage data of chromosome four of Drosophila melanogaster. Biol Zh. 1935;4:401–420.
-
- Carvalho AB, Clark AG. Y chromosome of D. pseudoobscura is not homologous to the ancestral Drosophila Y. Science. 2005;307:108–110. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

