PlantMetabolomics.org: a web portal for plant metabolomics experiments
- PMID: 20147492
- PMCID: PMC2850039
- DOI: 10.1104/pp.109.151027
PlantMetabolomics.org: a web portal for plant metabolomics experiments
Abstract
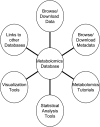
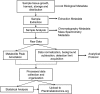
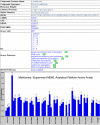
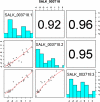
PlantMetabolomics.org (PM) is a web portal and database for exploring, visualizing, and downloading plant metabolomics data. Widespread public access to well-annotated metabolomics datasets is essential for establishing metabolomics as a functional genomics tool. PM integrates metabolomics data generated from different analytical platforms from multiple laboratories along with the key visualization tools such as ratio and error plots. Visualization tools can quickly show how one condition compares to another and which analytical platforms show the largest changes. The database tries to capture a complete annotation of the experiment metadata along with the metabolite abundance databased on the evolving Metabolomics Standards Initiative. PM can be used as a platform for deriving hypotheses by enabling metabolomic comparisons between genetically unique Arabidopsis (Arabidopsis thaliana) populations subjected to different environmental conditions. Each metabolite is linked to relevant experimental data and information from various annotation databases. The portal also provides detailed protocols and tutorials on conducting plant metabolomics experiments to promote metabolomics in the community. PM currently houses Arabidopsis metabolomics data generated by a consortium of laboratories utilizing metabolomics to help elucidate the functions of uncharacterized genes. PM is publicly available at http://www.plantmetabolomics.org.
Figures

References
-
- Altman DG, Bland JM. (1983) Measurement in medicine: the analysis of method comparison studies. Statistician 32: 307–317
-
- Bino RJ, Hall RD, Fiehn O, Kopka J, Saito K, Draper J, Nikolau BJ, Mendes P, Roessner-Tunali U, Beale MH, et al. (2004) Potential of metabolomics as a functional genomics tool. Trends Plant Sci 9: 418–425 - PubMed
-
- Cui Q, Lewis IA, Hegeman AD, Anderson ME, Li J, Schulte CF, Westler WM, Eghbalnia HR, Sussman MR, Markley JL. (2008) Metabolite identification via the Madison Metabolomics Consortium Database. Nat Biotechnol 26: 162–164 - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

