SUGAR-INSENSITIVE3, a RING E3 ligase, is a new player in plant sugar response
- PMID: 20147494
- PMCID: PMC2850021
- DOI: 10.1104/pp.109.150573
SUGAR-INSENSITIVE3, a RING E3 ligase, is a new player in plant sugar response
Abstract
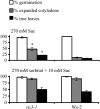
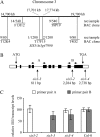
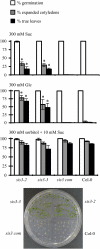
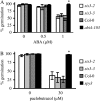
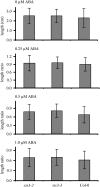
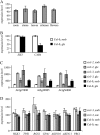
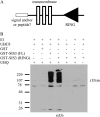
Sugars, such as sucrose and glucose, have been implicated in the regulation of diverse developmental events in plants and other organisms. We isolated an Arabidopsis (Arabidopsis thaliana) mutant, sugar-insensitive3 (sis3), that is resistant to the inhibitory effects of high concentrations of exogenous glucose and sucrose on early seedling development. In contrast to wild-type plants, sis3 mutants develop green, expanded cotyledons and true leaves when sown on medium containing high concentrations (e.g. 270 mm) of sucrose. Unlike some other sugar response mutants, sis3 exhibits wild-type responses to the inhibitory effects of abscisic acid and paclobutrazol, a gibberellic acid biosynthesis inhibitor, on seed germination. Map-based cloning revealed that SIS3 encodes a RING finger protein. Complementation of the sis3-2 mutant with a genomic SIS3 clone restored sugar sensitivity of sis3-2, confirming the identity of the SIS3 gene. Biochemical analyses demonstrated that SIS3 is functional in an in vitro ubiquitination assay and that the RING motif is sufficient for its activity. Our results indicate that SIS3 encodes a ubiquitin E3 ligase that is a positive regulator of sugar signaling during early seedling development.
Figures
Similar articles
-
SIS8, a putative mitogen-activated protein kinase kinase kinase, regulates sugar-resistant seedling development in Arabidopsis.Plant J. 2014 Feb;77(4):577-88. doi: 10.1111/tpj.12404. Epub 2014 Jan 23. Plant J. 2014. PMID: 24320620
-
The Arabidopsis sugar-insensitive mutants sis4 and sis5 are defective in abscisic acid synthesis and response.Plant J. 2000 Sep;23(5):587-96. doi: 10.1046/j.1365-313x.2000.00833.x. Plant J. 2000. PMID: 10972885
-
Identification, cloning and characterization of sis7 and sis10 sugar-insensitive mutants of Arabidopsis.BMC Plant Biol. 2008 Oct 14;8:104. doi: 10.1186/1471-2229-8-104. BMC Plant Biol. 2008. PMID: 18854047 Free PMC article.
-
The Arabidopsis RING finger E3 ligase RHA2a is a novel positive regulator of abscisic acid signaling during seed germination and early seedling development.Plant Physiol. 2009 May;150(1):463-81. doi: 10.1104/pp.109.135269. Epub 2009 Mar 13. Plant Physiol. 2009. PMID: 19286935 Free PMC article.
-
Interaction between sugar and abscisic acid signalling during early seedling development in Arabidopsis.Plant Mol Biol. 2008 May;67(1-2):151-67. doi: 10.1007/s11103-008-9308-6. Epub 2008 Feb 17. Plant Mol Biol. 2008. PMID: 18278579 Free PMC article.
Cited by
-
Transcriptome-Wide Analysis and Functional Verification of RING-Type Ubiquitin Ligase Involved in Tea Plant Stress Resistance.Front Plant Sci. 2021 Oct 21;12:733287. doi: 10.3389/fpls.2021.733287. eCollection 2021. Front Plant Sci. 2021. PMID: 34745167 Free PMC article.
-
Turning the Knobs: The Impact of Post-translational Modifications on Carbon Metabolism.Front Plant Sci. 2022 Jan 11;12:781508. doi: 10.3389/fpls.2021.781508. eCollection 2021. Front Plant Sci. 2022. PMID: 35087551 Free PMC article. Review.
-
The ABI4-induced Arabidopsis ANAC060 transcription factor attenuates ABA signaling and renders seedlings sugar insensitive when present in the nucleus.PLoS Genet. 2014 Mar 13;10(3):e1004213. doi: 10.1371/journal.pgen.1004213. eCollection 2014 Mar. PLoS Genet. 2014. PMID: 24625790 Free PMC article.
-
Arabidopsis Hexokinase-Like1 and Hexokinase1 form a critical node in mediating plant glucose and ethylene responses.Plant Physiol. 2012 Apr;158(4):1965-75. doi: 10.1104/pp.112.195636. Epub 2012 Feb 24. Plant Physiol. 2012. PMID: 22366209 Free PMC article.
-
A gene co-expression network predicts functional genes controlling the re-establishment of desiccation tolerance in germinated Arabidopsis thaliana seeds.Planta. 2015 Aug;242(2):435-49. doi: 10.1007/s00425-015-2283-7. Epub 2015 Mar 26. Planta. 2015. PMID: 25809152 Free PMC article.
References
-
- Baena-González E, Rolland F, Thevelein JM, Sheen J. (2007) A central integrator of transcription networks in plant stress and energy signalling. Nature 448: 938–942 - PubMed
-
- Bendtsen JD, Nielsen H, von Heijne G, Brunak S. (2004) Improved prediction of signal peptides: SignalP 3.0. J Mol Biol 340: 783–795 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

