Activating K-Ras mutations outwith 'hotspot' codons in sporadic colorectal tumours - implications for personalised cancer medicine
- PMID: 20147967
- PMCID: PMC2837563
- DOI: 10.1038/sj.bjc.6605534
Activating K-Ras mutations outwith 'hotspot' codons in sporadic colorectal tumours - implications for personalised cancer medicine
Abstract
Background: Response to EGFR-targeted therapies in colorectal cancer patients has been convincingly associated with Kirsten-Ras (K-Ras) mutation status. Current mandatory mutation testing for patient selection is limited to the K-Ras 'hotspot' codons 12 and 13.
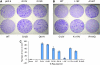
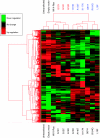
Methods: Colorectal tumours (n=106) were screened for additional K-Ras mutations, phenotypes compared in transformation and Ras GTPase activating assays and gene and pathway changes induced by individual K-Ras mutants identified by microarray analysis. Taqman-based gene copy number and FISH analyses were used to investigate K-Ras gene amplification.
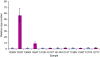
Results: Four additional K-Ras mutations (Leu(19)Phe (1 out of 106 tumours), Lys(117)Asn (1 out of 106), Ala(146)Thr (7 out of 106) and Arg(164)Gln (1 out of 106)) were identified. Lys(117)Asn and Ala(146)Thr had phenotypes similar to the hotspot mutations, whereas Leu(19)Phe had an attenuated phenotype and the Arg(164)Gln mutation was phenotypically equivalent to wt K-Ras. We additionally identified a new K-Ras gene amplification event, present in approximately 2% of tumours.
Conclusions: The identification of mutations outwith previously described hotspot codons increases the K-Ras mutation burden in colorectal tumours by one-third. Future mutation screening to facilitate optimal patient selection for treatment with EGFR-targeted therapies should therefore be extended to codon 146, and in addition should consider the unique molecular signatures associated with individual K-Ras mutations.
Figures

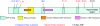

Comment in
-
Screening: A re-evaluation of KRAS mutational 'hotspots'.Nat Rev Clin Oncol. 2010 May;7(5):242. doi: 10.1038/nrclinonc.2010.54. Nat Rev Clin Oncol. 2010. PMID: 20432530 No abstract available.
Similar articles
-
Epidermal growth factor receptor (EGFR) status and K-Ras mutations in colorectal cancer.Ann Oncol. 2008 Dec;19(12):2033-8. doi: 10.1093/annonc/mdn416. Epub 2008 Jul 15. Ann Oncol. 2008. PMID: 18632722 Free PMC article.
-
Predominance of G to A codon 12 mutation K-ras gene in Dukes' B colorectal cancer.Singapore Med J. 2012 Jan;53(1):26-31. Singapore Med J. 2012. PMID: 22252179
-
Mutations in APC, CTNNB1 and K-ras genes and expression of hMLH1 in sporadic colorectal carcinomas from the Netherlands Cohort Study.BMC Cancer. 2005 Dec 15;5:160. doi: 10.1186/1471-2407-5-160. BMC Cancer. 2005. PMID: 16356174 Free PMC article.
-
Ras oncogene and p53 gene hotspot mutations in colorectal cancers.J Gastroenterol Hepatol. 1995 Mar-Apr;10(2):119-24. doi: 10.1111/j.1440-1746.1995.tb01064.x. J Gastroenterol Hepatol. 1995. PMID: 7787154
-
The ideal reporting of RAS testing in colorectal adenocarcinoma: a pathologists' perspective.Pathologica. 2023 Jun 14;115(3):137-47. doi: 10.32074/1591-951X-895. Online ahead of print. Pathologica. 2023. PMID: 37314870 Free PMC article. Review.
Cited by
-
Intraductal Transplantation Models of Human Pancreatic Ductal Adenocarcinoma Reveal Progressive Transition of Molecular Subtypes.Cancer Discov. 2020 Oct;10(10):1566-1589. doi: 10.1158/2159-8290.CD-20-0133. Epub 2020 Jul 23. Cancer Discov. 2020. PMID: 32703770 Free PMC article.
-
Functional and biological heterogeneity of KRASQ61 mutations.Sci Signal. 2022 Aug 9;15(746):eabn2694. doi: 10.1126/scisignal.abn2694. Epub 2022 Aug 9. Sci Signal. 2022. PMID: 35944066 Free PMC article.
-
A Cross-Sectional Study for Evaluation of KRAS and BRAF Mutations by Reverse Dot Blot, PCR-RFLP, and Allele-Specific PCR Methods Among Patients with Colorectal Cancer.Avicenna J Med Biotechnol. 2021 Oct-Dec;13(4):183-191. Avicenna J Med Biotechnol. 2021. PMID: 34900144 Free PMC article.
-
Mutations of p53 and K-ras correlate TF expression in human colorectal carcinomas: TF downregulation as a marker of poor prognosis.Int J Colorectal Dis. 2011 May;26(5):593-601. doi: 10.1007/s00384-011-1164-1. Epub 2011 Mar 15. Int J Colorectal Dis. 2011. PMID: 21404058
-
Review: Precision medicine and driver mutations: Computational methods, functional assays and conformational principles for interpreting cancer drivers.PLoS Comput Biol. 2019 Mar 28;15(3):e1006658. doi: 10.1371/journal.pcbi.1006658. eCollection 2019 Mar. PLoS Comput Biol. 2019. PMID: 30921324 Free PMC article. Review.
References
-
- Akagi K, Uchibori R, Yamaguchi K, Kurosawa K, Tanaka Y, Kozu T (2007) Characterization of a novel oncogenic K-ras mutation in colon cancer. Biochem Biophys Res Commun 352: 728–732 - PubMed
-
- Andreyev HJ, Norman AR, Cunningham D, Oates J, Dix BR, Iacopetta BJ, Young J, Walsh T, Ward R, Hawkins N, Beranek M, Jandik P, Benamouzig R, Jullian E, Laurent-Puig P, Olschwang S, Muller O, Hoffmann I, Rabes HM, Zietz C, Troungos C, Valavanis C, Yuen ST, Ho JW, Croke CT, O'Donoghue DP, Giaretti W, Rapallo A, Russo A, Bazan V, Tanaka M, Omura K, Azuma T, Ohkusa T, Fujimori T, Ono Y, Pauly M, Faber C, Glaesener R, de Goeij AF, Arends JW, Andersen SN, Lovig T, Breivik J, Gaudernack G, Clausen OP, De Angelis PD, Meling GI, Rognum TO, Smith R, Goh HS, Font A, Rosell R, Sun XF, Zhang H, Benhattar J, Losi L, Lee JQ, Wang ST, Clarke PA, Bell S, Quirke P, Bubb VJ, Piris J, Cruickshank NR, Morton D, Fox JC, Al-Mulla F, Lees N, Hall CN, Snary D, Wilkinson K, Dillon D, Costa J, Pricolo VE, Finkelstein SD, Thebo JS, Senagore AJ, Halter SA, Wadler S, Malik S, Krtolica K, Urosevic N (2001) Kirsten ras mutations in patients with colorectal cancer: the ‘RASCAL II’ study. Br J Cancer 85: 692–696 - PMC - PubMed
-
- Barbacid M (1990) Ras oncogenes: their role in neoplasia. Eur J Clin Invest 20: 225–235 - PubMed
-
- Benvenuti S, Sartore-Bianchi A, Di Nicolantonio F, Zanon C, Moroni M, Veronese S, Siena S, Bardelli A (2007) Oncogenic activation of the RAS/RAF signaling pathway impairs the response of metastatic colorectal cancers to anti-epidermal growth factor receptor antibody therapies. Cancer Res 67: 2643–2648 - PubMed
-
- Bloethner S, Chen B, Hemminki K, Muller-Berghaus J, Ugurel S, Schadendorf D, Kumar R (2005) Effect of common B-RAF and N-RAS mutations on global gene expression in melanoma cell lines. Carcinogenesis 26: 1224–1232 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

