The application of restriction landmark genome scanning method for surveillance of non-mendelian inheritance in f(1) hybrids
- PMID: 20148066
- PMCID: PMC2817499
- DOI: 10.1155/2009/245927
The application of restriction landmark genome scanning method for surveillance of non-mendelian inheritance in f(1) hybrids
Abstract
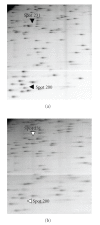
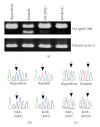
We analyzed inheritance of DNA methylation in reciprocal F(1) hybrids (subsp. japonica cv. Nipponbare x subsp. indica cv. Kasalath) of rice (Oryza sativa L.) using restriction landmark genome scanning (RLGS), and detected differing RLGS spots between the parents and reciprocal F(1) hybrids. MspI/HpaII restriction sites in the DNA from these different spots were suspected to be heterozygously methylated in the Nipponbare parent. These spots segregated in F(1) plants, but did not segregate in selfed progeny of Nipponbare, showing non-Mendelian inheritance of the methylation status. As a result of RT-PCR and sequencing, a specific allele of the gene nearest to the methylated sites was expressed in reciprocal F(1) plants, showing evidence of biased allelic expression. These results show the applicability of RLGS for scanning of non-Mendelian inheritance of DNA methylation and biased allelic expression.
Figures

Similar articles
-
Inheritance and alteration of genome methylation in F1 hybrid rice.Electrophoresis. 2008 Oct;29(19):4088-95. doi: 10.1002/elps.200700784. Electrophoresis. 2008. PMID: 18958879
-
Restriction landmark genome scanning.Methods Mol Biol. 2011;791:101-12. doi: 10.1007/978-1-61779-316-5_8. Methods Mol Biol. 2011. PMID: 21913074
-
Inheritance of Oryza sativa endornavirus in F1 and F2 hybrids between japonica and indica rice.Genes Genet Syst. 2003 Jun;78(3):229-34. doi: 10.1266/ggs.78.229. Genes Genet Syst. 2003. PMID: 12893964
-
Epigenetics: application of virtual image restriction landmark genomic scanning (Vi-RLGS).FEBS J. 2008 Apr;275(8):1608-16. doi: 10.1111/j.1742-4658.2008.06329.x. Epub 2008 Mar 7. FEBS J. 2008. PMID: 18331348 Review.
-
Restriction landmark genomic scanning for DNA methylation in cancer: past, present, and future applications.Anal Biochem. 2002 Aug 15;307(2):191-201. doi: 10.1016/s0003-2697(02)00033-7. Anal Biochem. 2002. PMID: 12202234 Review.
Cited by
-
Identification of Imprinted Genes Based on Homology: An Example of Fragaria vesca.Genes (Basel). 2021 Mar 8;12(3):380. doi: 10.3390/genes12030380. Genes (Basel). 2021. PMID: 33800118 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

