Coupling Constant pH Molecular Dynamics with Accelerated Molecular Dynamics
- PMID: 20148176
- PMCID: PMC2817915
- DOI: 10.1021/ct9005294
Coupling Constant pH Molecular Dynamics with Accelerated Molecular Dynamics
Abstract
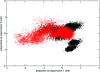
An extension of the constant pH method originally implemented by Mongan et al. (J. Comput. Chem.2004, 25, 2038-2048) is proposed in this study. This adapted version of the method couples the constant pH methodology with the enhanced sampling technique of accelerated molecular dynamics, in an attempt to overcome the sampling issues encountered with current standard constant pH molecular dynamics methods. Although good results were reported by Mongan et al. on application of the standard method to the hen egg-white lysozyme (HEWL) system, residues which possess strong interactions with neighboring groups tend to converge slowly, resulting in the reported inconsistencies for predicted pK(a) values, as highlighted by the authors. The application of the coupled method described in this study to the HEWL system displays improvements over the standard version of the method, with the improved sampling leading to faster convergence and producing pK(a) values in closer agreement to those obtained experimentally for the more slowly converging residues.
Figures




References
-
- Baptista A. M.; Teixeira V. H.; Soares C. M. J. Chem. Phys. 2002, 117, 4184–4200.
-
- Dlugosz M.; Antosiewicz J. M.; Robertson A. D. Phys. Rev. E 2004, 69, 021915.1–021915.10. - PubMed
-
- Dlugosz M.; Antosiewicz J. M. Chem. Phys. 2004, 302, 161–170.
-
- Mongan J.; Case D. A.; McCammon J. A. J. Comput. Chem. 2004, 25, 2038–2048. - PubMed
-
- Baptista A. M.; Martel P. J.; Petersen S. B. Proteins: Struct., Funct., Genet. 1997, 27, 523–544. - PubMed

