Suprachiasmatic nucleus: cell autonomy and network properties
- PMID: 20148688
- PMCID: PMC3758475
- DOI: 10.1146/annurev-physiol-021909-135919
Suprachiasmatic nucleus: cell autonomy and network properties
Abstract
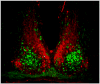
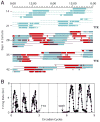
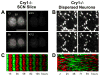
The suprachiasmatic nucleus (SCN) is the primary circadian pacemaker in mammals. Individual SCN neurons in dispersed culture can generate independent circadian oscillations of clock gene expression and neuronal firing. However, SCN rhythmicity depends on sufficient membrane depolarization and levels of intracellular calcium and cAMP. In the intact SCN, cellular oscillations are synchronized and reinforced by rhythmic synaptic input from other cells, resulting in a reproducible topographic pattern of distinct phases and amplitudes specified by SCN circuit organization. The SCN network synchronizes its component cellular oscillators, reinforces their oscillations, responds to light input by altering their phase distribution, increases their robustness to genetic perturbations, and enhances their precision. Thus, even though individual SCN neurons can be cell-autonomous circadian oscillators, neuronal network properties are integral to normal function of the SCN.
Figures

References
-
- Ueda HR, et al. System-level identification of transcriptional circuits underlying mammalian circadian clocks. Nat Genet. 2005;37:187–92. - PubMed
-
- Klein DC, Moore RY, Reppert SM, editors. Suprachiasmatic Nucleus: The Mind’s Clock. New York: Oxford University Press; 1991.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

