A comparison of selected quantitative trait loci associated with alcohol use phenotypes in humans and mouse models
- PMID: 20148779
- PMCID: PMC2848508
- DOI: 10.1111/j.1369-1600.2009.00195.x
A comparison of selected quantitative trait loci associated with alcohol use phenotypes in humans and mouse models
Abstract
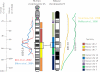
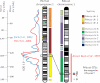
Evidence for genetic linkage to alcohol and other substance dependence phenotypes in areas of the human and mouse genome have now been reported with some consistency across studies. However, the question remains as to whether the genes that underlie the alcohol-related behaviors seen in mice are the same as those that underlie the behaviors observed in human alcoholics. The aims of the current set of analyses were to identify a small set of alcohol-related phenotypes in human and in mouse by which to compare quantitative trait locus (QTL) data between the species using syntenic mapping. These analyses identified that QTLs for alcohol consumption and acute and chronic alcohol withdrawal on distal mouse chromosome 1 are syntenic to a region on human chromosome 1q where a number of studies have identified QTLs for alcohol-related phenotypes. Additionally, a QTL on human chromosome 15 for alcohol dependence severity/withdrawal identified in two human studies was found to be largely syntenic with a region on mouse chromosome 9, where two groups have found QTLs for alcohol preference. In both of these cases, while the QTLs were found to be syntenic, the exact phenotypes between humans and mice did not necessarily overlap. These studies demonstrate how this technique might be useful in the search for genes underlying alcohol-related phenotypes in multiple species. However, these findings also suggest that trying to match exact phenotypes in humans and mice may not be necessary or even optimal for determining whether similar genes influence a range of alcohol-related behaviors between the two species.
Figures
Similar articles
-
Mapping a locus for alcohol physical dependence and associated withdrawal to a 1.1 Mb interval of mouse chromosome 1 syntenic with human chromosome 1q23.2-23.3.Genes Brain Behav. 2008 Jul;7(5):560-7. doi: 10.1111/j.1601-183X.2008.00391.x. Genes Brain Behav. 2008. PMID: 18363856
-
Identifying quantitative trait loci (QTLs) and genes (QTGs) for alcohol-related phenotypes in mice.Int Rev Neurobiol. 2010;91:173-204. doi: 10.1016/S0074-7742(10)91006-4. Int Rev Neurobiol. 2010. PMID: 20813243 Review.
-
Mapping murine loci for physical dependence on ethanol.Psychopharmacology (Berl). 2002 Apr;160(4):398-407. doi: 10.1007/s00213-001-0988-8. Epub 2002 Feb 15. Psychopharmacology (Berl). 2002. PMID: 11919667
-
Quantitative trait loci involved in genetic predisposition to acute alcohol withdrawal in mice.J Neurosci. 1997 May 15;17(10):3946-55. doi: 10.1523/JNEUROSCI.17-10-03946.1997. J Neurosci. 1997. PMID: 9133412 Free PMC article.
-
Strategies for mapping and identifying quantitative trait loci specifying behavioral responses to alcohol.Alcohol Clin Exp Res. 1995 Aug;19(4):795-801. doi: 10.1111/j.1530-0277.1995.tb00949.x. Alcohol Clin Exp Res. 1995. PMID: 7485822 Review.
Cited by
-
Fyn-dependent gene networks in acute ethanol sensitivity.PLoS One. 2013 Nov 29;8(11):e82435. doi: 10.1371/journal.pone.0082435. eCollection 2013. PLoS One. 2013. PMID: 24312422 Free PMC article.
-
Distinct Roles for Two Chromosome 1 Loci in Ethanol Withdrawal, Consumption, and Conditioned Place Preference.Front Genet. 2018 Aug 27;9:323. doi: 10.3389/fgene.2018.00323. eCollection 2018. Front Genet. 2018. PMID: 30210527 Free PMC article.
-
Modeling the diagnostic criteria for alcohol dependence with genetic animal models.Curr Top Behav Neurosci. 2013;13:187-221. doi: 10.1007/7854_2011_162. Curr Top Behav Neurosci. 2013. PMID: 21910077 Free PMC article. Review.
-
The multiple PDZ domain protein Mpdz/MUPP1 regulates opioid tolerance and opioid-induced hyperalgesia.BMC Genomics. 2016 Apr 29;17:313. doi: 10.1186/s12864-016-2634-1. BMC Genomics. 2016. PMID: 27129385 Free PMC article.
-
Expression, covariation, and genetic regulation of miRNA Biogenesis genes in brain supports their role in addiction, psychiatric disorders, and disease.Front Genet. 2013 Jul 5;4:126. doi: 10.3389/fgene.2013.00126. eCollection 2013. Front Genet. 2013. PMID: 23847651 Free PMC article.
References
-
- Agrawal A, Pergadia ML, Saccone SF, Lynskey MT, Wang JC, Martin NG, Statham D, Henders A, Campbell M, Garcia R, Broms U, Todd RD, Goate AM, Rice J, Kaprio J, Heath AC, Montgomery GW, Madden PA. An autosomal linkage scan for cannabis use disorders in the nicotine addiction genetics project. Arch Gen Psychiatry. 2008;65:713–721. - PubMed
-
- Aragaki C, Quiaoit F, Hsu L, Zhao LP. Mapping alcoholism genes using linkage/linkage disequilibrium analysis. Genet Epidemiol. 1999;17(Suppl 1):S43–S48. - PubMed
-
- Barr CS, Goldman D. Non-human primate models of inheritance vulnerability to alcohol use disorders. Addict Biol. 2006;11:374–385. - PubMed
-
- Becker HC. The alcohol withdrawal “kindling” phenomenon: clinical and experimental findings. Alcohol Clin Exp Res. 1996;20:121A–124A. - PubMed
-
- Belknap JK, Laursen SE, Crabbe JC. Ethanol and nitrous oxide produce withdrawal-induced convulsions by similar mechanisms in mice. Life Sci. 1987;41:2033–2040. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- AA011114/AA/NIAAA NIH HHS/United States
- R01 AA011114/AA/NIAAA NIH HHS/United States
- P50 AA010760/AA/NIAAA NIH HHS/United States
- R01 AA010201/AA/NIAAA NIH HHS/United States
- R29 AA011114/AA/NIAAA NIH HHS/United States
- K02 AA018755/AA/NIAAA NIH HHS/United States
- R01 AA012714/AA/NIAAA NIH HHS/United States
- R37 AA010201/AA/NIAAA NIH HHS/United States
- AA010201/AA/NIAAA NIH HHS/United States
- R01 DA005228/DA/NIDA NIH HHS/United States
- R01 AA006059/AA/NIAAA NIH HHS/United States
- DA005228/DA/NIDA NIH HHS/United States
- P60 AA010760/AA/NIAAA NIH HHS/United States
- AA 006059/AA/NIAAA NIH HHS/United States
- U54 RR025204/RR/NCRR NIH HHS/United States
- AA12714/AA/NIAAA NIH HHS/United States
- AA010760/AA/NIAAA NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical

