PLX4032, a selective BRAF(V600E) kinase inhibitor, activates the ERK pathway and enhances cell migration and proliferation of BRAF melanoma cells
- PMID: 20149136
- PMCID: PMC2848976
- DOI: 10.1111/j.1755-148X.2010.00685.x
PLX4032, a selective BRAF(V600E) kinase inhibitor, activates the ERK pathway and enhances cell migration and proliferation of BRAF melanoma cells
Erratum in
- Pigment Cell Melanoma Res. 2012 May;25(3):402
Abstract
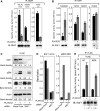
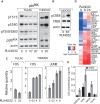
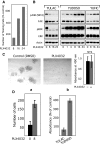
BRAF(V600E/K) is a frequent mutationally active tumor-specific kinase in melanomas that is currently targeted for therapy by the specific inhibitor PLX4032. Our studies with melanoma tumor cells that are BRAF(V600E/K) and BRAF(WT) showed that, paradoxically, while PLX4032 inhibited ERK1/2 in the highly sensitive BRAF(V600E/K), it activated the pathway in the resistant BRAF(WT) cells, via RAF1 activation, regardless of the status of mutations in NRAS or PTEN. The persistently active ERK1/2 triggered downstream effectors in BRAF(WT) melanoma cells and induced changes in the expression of a wide-spectrum of genes associated with cell cycle control. Furthermore, PLX4032 increased the rate of proliferation of growth factor-dependent NRAS Q61L mutant primary melanoma cells, reduced cell adherence and increased mobility of cells from advanced lesions. The results suggest that the drug can confer an advantage to BRAF(WT) primary and metastatic tumor cells in vivo and provide markers for monitoring clinical responses.
Figures
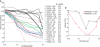


References
-
- Baccarini M. Second nature: biological functions of the Raf-1 “kinase”. FEBS Lett. 2005;579:3271–3277. - PubMed
-
- Bild AH, Yao G, Chang JT, et al. Oncogenic pathway signatures in human cancers as a guide to targeted therapies. Nature. 2006;439:353–357. - PubMed
-
- Böhm M, Moellmann G, Cheng E, Alvarez-Franco M, Wagner S, Sassone-Corsi P, Halaban R. Identification of p90RSK as the probable CREB-Ser133 kinase in human melanocytes. Cell Growth Differ. 1995;6:291–302. - PubMed
-
- Craig EA, Stevens MV, Vaillancourt RR, Camenisch TD. MAP3Ks as central regulators of cell fate during development. Dev. Dyn. 2008;237:3102–3114. - PubMed
-
- Curtin JA, Busam K, Pinkel D, Bastian BC. Somatic activation of KIT in distinct subtypes of melanoma. J. Clin. Oncol. 2006;24:4340–4346. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

