Host genes associated with HIV/AIDS: advances in gene discovery
- PMID: 20149939
- PMCID: PMC3792714
- DOI: 10.1016/j.tig.2010.01.002
Host genes associated with HIV/AIDS: advances in gene discovery
Abstract
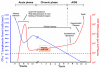
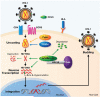
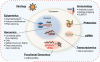
Twenty-five years after the discovery of HIV as the cause of AIDS there is still no effective vaccine and no cure for this disease. HIV susceptibility shows a substantial degree of individual heterogeneity, much of which can be conferred by host genetic variation. In an effort to discover host factors required for HIV replication, identify crucial pathogenic pathways, and reveal the full armament of host defenses, there has been a shift from candidate-gene studies to unbiased genome-wide genetic and functional studies. However, the number of securely identified host factors involved in HIV disease remains small, explaining only approximately 15-20% of the observed heterogeneity - most of which is attributable to human lymphocyte antigen (HLA) variants. Multidisciplinary approaches integrating genetic epidemiology to systems biology will be required to fully understand virus-host interactions to effectively combat HIV/AIDS.
Copyright 2010 Elsevier Ltd. All rights reserved.
Figures


References
-
- O’Brien SJ, Nelson GW. Human genes that limit AIDS. Nature genetics. 2004;36:565–574. - PubMed
-
- Dean M, et al. Genetic restriction of HIV-1 infection and progression to AIDS by a deletion allele of the CKR5 structural gene. Hemophilia Growth and Development Study, Multicenter AIDS Cohort Study, Multicenter Hemophilia Cohort Study, San Francisco City Cohort, ALIVE Study. Science. 1996;273:1856–1862. - PubMed
-
- Hutter G, et al. Long-term control of HIV by CCR5 Delta32/Delta32 stem-cell transplantation. N Engl J Med. 2009;360:692–698. - PubMed
-
- Shacklett BL. Understanding the “lucky few”: the conundrum of HIV-exposed, seronegative individuals. Current HIV/AIDS reports. 2006;3:26–31. - PubMed
-
- Walker BD. Elite control of HIV Infection: implications for vaccines and treatment. Top HIV Med. 2007;15:134–136. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

