Analysis of the liver mitochondrial proteome in response to ethanol and S-adenosylmethionine treatments: novel molecular targets of disease and hepatoprotection
- PMID: 20150243
- PMCID: PMC2867419
- DOI: 10.1152/ajpgi.00332.2009
Analysis of the liver mitochondrial proteome in response to ethanol and S-adenosylmethionine treatments: novel molecular targets of disease and hepatoprotection
Abstract
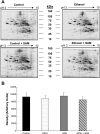
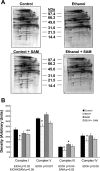
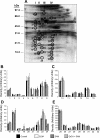
S-adenosylmethionine (SAM) minimizes alcohol hepatotoxicity; however, the molecular mechanisms responsible for SAM hepatoprotection remain unknown. Herein, we use proteomics to determine whether the hepatoprotective action of SAM against early-stage alcoholic liver disease is linked to alterations in the mitochondrial proteome. For this, male rats were fed control or ethanol-containing liquid diets +/- SAM and liver mitochondria were prepared for proteomic analysis. Two-dimensional isoelectric focusing (2D IEF/SDS-PAGE) and blue native gel electrophoresis (BN-PAGE) were used to determine changes in matrix and oxidative phosphorylation (OxPhos) proteins, respectively. SAM coadministration minimized alcohol-dependent inflammation and preserved mitochondrial respiration. SAM supplementation preserved liver SAM levels in ethanol-fed rats; however, mitochondrial SAM levels were increased by ethanol and SAM treatments. With use of 2D IEF/SDS-PAGE, 30 proteins showed significant changes in abundance in response to ethanol, SAM, or both. Classes of proteins affected by ethanol and SAM treatments were chaperones, beta oxidation proteins, sulfur metabolism proteins, and dehydrogenase enzymes involved in methionine, glycine, and choline metabolism. BN-PAGE revealed novel changes in the levels of 19 OxPhos proteins in response to ethanol, SAM, or both. Ethanol- and SAM-dependent alterations in the proteome were not linked to corresponding changes in gene expression. In conclusion, ethanol and SAM treatment led to multiple changes in the liver mitochondrial proteome. The protective effects of SAM against alcohol toxicity are mediated, in part, through maintenance of proteins involved in key mitochondrial energy conserving and biosynthetic pathways. This study demonstrates that SAM may be a promising candidate for treatment of alcoholic liver disease.
Figures



References
-
- Alnouti Y, Klaassen CD. Tissue distribution, ontogeny, and regulation of aldehyde dehydrogenase (Aldh) enzymes mRNA by prototypical microsomal enzyme inducers in mice. Toxicol Sci 101: 51–64, 2008 - PubMed
-
- Andringa K, King A, Bailey S. Blue native-gel electrophoresis proteomics. Methods Mol Biol 519: 241–258, 2009 - PubMed
-
- Arai M, Leo MA, Nakano M, Gordon ER, Lieber CS. Biochemical and morphological alterations of baboon hepatic mitochondria after chronic ethanol consumption. Hepatology 4: 165–174, 1984 - PubMed
-
- Bailey SM, Cunningham CC. Contribution of mitochondria to oxidative stress associated with alcoholic liver disease. Free Radic Biol Med 32: 11–16, 2002 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

