Accurate detection and genotyping of SNPs utilizing population sequencing data
- PMID: 20150320
- PMCID: PMC2847757
- DOI: 10.1101/gr.100040.109
Accurate detection and genotyping of SNPs utilizing population sequencing data
Abstract
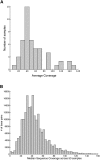
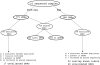
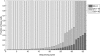
Next-generation sequencing technologies have made it possible to sequence targeted regions of the human genome in hundreds of individuals. Deep sequencing represents a powerful approach for the discovery of the complete spectrum of DNA sequence variants in functionally important genomic intervals. Current methods for single nucleotide polymorphism (SNP) detection are designed to detect SNPs from single individual sequence data sets. Here, we describe a novel method SNIP-Seq (single nucleotide polymorphism identification from population sequence data) that leverages sequence data from a population of individuals to detect SNPs and assign genotypes to individuals. To evaluate our method, we utilized sequence data from a 200-kilobase (kb) region on chromosome 9p21 of the human genome. This region was sequenced in 48 individuals (five sequenced in duplicate) using the Illumina GA platform. Using this data set, we demonstrate that our method is highly accurate for detecting variants and can filter out false SNPs that are attributable to sequencing errors. The concordance of sequencing-based genotype assignments between duplicate samples was 98.8%. The 200-kb region was independently sequenced to a high depth of coverage using two sequence pools containing the 48 individuals. Many of the novel SNPs identified by SNIP-Seq from the individual sequencing were validated by the pooled sequencing data and were subsequently confirmed by Sanger sequencing. We estimate that SNIP-Seq achieves a low false-positive rate of approximately 2%, improving upon the higher false-positive rate for existing methods that do not utilize population sequence data. Collectively, these results suggest that analysis of population sequencing data is a powerful approach for the accurate detection of SNPs and the assignment of genotypes to individual samples.
Figures

References
-
- Cohen JC, Kiss RS, Pertsemlidis A, Marcel YL, McPherson R, Hobbs HH. Multiple rare alleles contribute to low plasma levels of HDL cholesterol. Science. 2004;305:869–872. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
