Role of miR-34c microRNA in the late steps of spermatogenesis
- PMID: 20150330
- PMCID: PMC2844620
- DOI: 10.1261/rna.1963810
Role of miR-34c microRNA in the late steps of spermatogenesis
Abstract
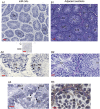
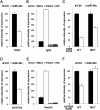
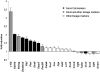
Spermatogenesis is a cyclic process in which diploid spermatogonia differentiate into haploid spermatozoa. This process is highly regulated, notably at the post-transcriptional level. MicroRNAs (miRNAs), single-stranded noncoding RNA molecules of about 20-25 nucleotides, are implicated in the regulation of many important biological pathways such as proliferation, apoptosis, and differentiation. We wondered whether miRNAs could play a role during spermatogenesis. The miRNA expression repertoire was tested in germ cells, and we present data showing that miR-34c was highly expressed only in these cells. Furthermore, our findings indicate that in male gonads, miR-34c expression is largely p53 independent in contrast to previous results showing a direct link in somatic cells between the miR-34 family and this tumor suppressor protein. In order to identify target genes involved in germinal lineage differentiation, we overexpressed miR-34c in HeLa cells, analyzed the transcriptome of these modified cells, and noticed a shift of the expression profile toward the germinal lineage. Recently, it has been shown that exogenous expression of Ddx4/Vasa in embryonic chicken stem cells (cESC) induces cESC reprogramming toward a germ cell fate. When we simultaneously expressed miR-34c in such cells, we could detect an up-regulation of germ cell-specific genes whereas the expression of other lineage specific markers remained unchanged. These data suggest that miR-34c could play a role by enhancing the germinal phenotype of cells already committed to this lineage.
Figures




Similar articles
-
MIR-34c regulates mouse embryonic stem cells differentiation into male germ-like cells through RARg.Cell Biochem Funct. 2012 Dec;30(8):623-32. doi: 10.1002/cbf.2922. Epub 2012 Oct 24. Cell Biochem Funct. 2012. PMID: 23097316
-
Expression of miR-34c in response to overexpression of Boule and Stra8 in dairy goat male germ line stem cells (mGSCs).Cell Biochem Funct. 2013 Jun;31(4):281-8. doi: 10.1002/cbf.2970. Epub 2013 Mar 19. Cell Biochem Funct. 2013. PMID: 23508548
-
miR-34c works downstream of p53 leading to dairy goat male germline stem-cell (mGSCs) apoptosis.Cell Prolif. 2013 Apr;46(2):223-31. doi: 10.1111/cpr.12013. Cell Prolif. 2013. PMID: 23510477 Free PMC article.
-
MicroRNAs and spermatogenesis.Fertil Steril. 2014 Jun;101(6):1552-62. doi: 10.1016/j.fertnstert.2014.04.025. Fertil Steril. 2014. PMID: 24882619 Review.
-
microRNA as an Important Mediator in the Regulation of Male Gallus gallus domesticus Reproduction: Current State of the Problem.Int J Mol Sci. 2024 Dec 26;26(1):112. doi: 10.3390/ijms26010112. Int J Mol Sci. 2024. PMID: 39795968 Free PMC article. Review.
Cited by
-
Intact p53-dependent responses in miR-34-deficient mice.PLoS Genet. 2012;8(7):e1002797. doi: 10.1371/journal.pgen.1002797. Epub 2012 Jul 26. PLoS Genet. 2012. PMID: 22844244 Free PMC article.
-
MicroRNA profiles in a monkey testicular injury model induced by testicular hyperthermia.J Appl Toxicol. 2016 Dec;36(12):1614-1621. doi: 10.1002/jat.3326. Epub 2016 Apr 12. J Appl Toxicol. 2016. PMID: 27071960 Free PMC article.
-
Next Generation Sequencing Analysis Reveals Segmental Patterns of microRNA Expression in Mouse Epididymal Epithelial Cells.PLoS One. 2015 Aug 13;10(8):e0135605. doi: 10.1371/journal.pone.0135605. eCollection 2015. PLoS One. 2015. PMID: 26270822 Free PMC article.
-
Inflammatory Modulation of miR-155 Inhibits Doxorubicin-Induced Testicular Dysfunction via SIRT1/FOXO1 Pathway: Insight into the Role of Acacetin and Bacillus cereus Protease.Appl Biochem Biotechnol. 2022 Nov;194(11):5196-5219. doi: 10.1007/s12010-022-03992-8. Epub 2022 Jun 18. Appl Biochem Biotechnol. 2022. PMID: 35715546 Free PMC article.
-
A survey of small RNAs in human sperm.Hum Reprod. 2011 Dec;26(12):3401-12. doi: 10.1093/humrep/der329. Epub 2011 Oct 11. Hum Reprod. 2011. PMID: 21989093 Free PMC article.
References
-
- Ambros V. The evolution of our thinking about microRNAs. Nat Med. 2008;14:1036–1040. - PubMed
-
- Bommer GT, Gerin I, Feng Y, Kaczorowski AJ, Kuick R, Love RE, Zhai Y, Giordano TJ, Qin ZS, Moore BB, et al. p53-mediated activation of miRNA34 candidate tumor-suppressor genes. Curr Biol. 2007;17:1298–1307. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
