Identification of novel oxidized protein substrates and physiological partners of the mitochondrial ATP-dependent Lon-like protease Pim1
- PMID: 20150421
- PMCID: PMC2857023
- DOI: 10.1074/jbc.M109.065425
Identification of novel oxidized protein substrates and physiological partners of the mitochondrial ATP-dependent Lon-like protease Pim1
Abstract
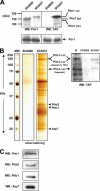
ATP-dependent proteases are currently emerging as key regulators of mitochondrial functions. Among these proteolytic systems, Pim1, a Lon-like serine protease in Saccharomyces cerevisiae, is involved in the control of selective protein turnover in the mitochondrial matrix. In the absence of Pim1, yeast cells have been shown to accumulate electron-dense inclusion bodies in the matrix space, to lose integrity of mitochondrial genome, and to be respiration-deficient. Because of the severity of phenotypes associated with the depletion of Pim1, this protease appears to be an essential component of the protein quality control machinery in mitochondria and to exert crucial functions during the biogenesis of this organelle. Nevertheless, its physiological substrates and partners are not fully characterized. Therefore, we used the combination of different proteomic techniques to assess the nature of oxidized protein substrates and physiological partners of Pim1 protease under non-repressing growth conditions. The results presented here supply evidence that Pim1-mediated proteolysis is required for elimination of oxidatively damaged proteins in mitochondria.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

