Structure of sterol aliphatic chains affects yeast cell shape and cell fusion during mating
- PMID: 20150508
- PMCID: PMC2840153
- DOI: 10.1073/pnas.0914094107
Structure of sterol aliphatic chains affects yeast cell shape and cell fusion during mating
Abstract
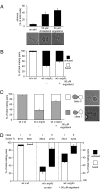
Under mating conditions, yeast cells adopt a characteristic pear-shaped morphology, called a "shmoo," as they project a cell extension toward their mating partners. Mating partners make contact at their shmoo tips, dissolve the intervening cell wall, and fuse their plasma membranes. We identified mutations in ERG4, encoding the enzyme that catalyzes the last step of ergosterol biosynthesis, that impair both shmoo formation and cell fusion. Upon pheromone treatment, erg4Delta mutants polarized growth, lipids, and proteins involved in mating but did not form properly shaped shmoos and fused with low efficiency. Supplementation with ergosterol partially suppressed the shmooing defect but not the cell fusion defect. By contrast, removal of the Erg4 substrate ergosta-5,7,22,24(28)-tetraenol, which accumulates in erg4Delta mutant cells and contains an extra double bond in the aliphatic chain of the sterol, restored both shmooing and cell fusion to wild-type levels. Thus, a two-atom change in the aliphatic moiety of ergosterol is sufficient to obstruct cell shape remodeling and cell fusion.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Chen EH, Grote E, Mohler W, Vignery A. Cell-cell fusion. FEBS Lett. 2007;581:2181–2193. - PubMed
-
- Ydenberg CA, Rose MD. Yeast mating: a model system for studying cell and nuclear fusion. Methods Mol Biol. 2008;475:3–20. - PubMed
-
- Herskowitz I. MAP kinase pathways in yeast: for mating and more. Cell. 1995;80:187–197. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

