A role for Sp1 in transcriptional regulation of phosphatidylethanolamine N-methyltransferase in liver and 3T3-L1 adipocytes
- PMID: 20150657
- PMCID: PMC2852925
- DOI: 10.1074/jbc.M110.109843
A role for Sp1 in transcriptional regulation of phosphatidylethanolamine N-methyltransferase in liver and 3T3-L1 adipocytes
Abstract
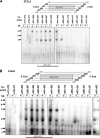
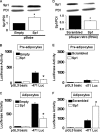
Phosphatidylcholine is made in all nucleated mammalian cells via the CDP-choline pathway. Another major pathway for phosphatidylcholine biosynthesis in liver is catalyzed by phosphatidylethanolamine N-methyltransferase (PEMT). We have now identified 3T3-L1 adipocytes as a cell culture model that expresses PEMT endogenously. We have found that PEMT mRNA and protein levels increased dramatically in 3T3-L1 cells upon differentiation to adipocytes. 5'-Deletion analysis of the PEMT promoter-luciferase constructs stably expressed in 3T3-L1 adipocytes identified a regulatory region between -471 and -371 bp (relative to the transcriptional start site). Competitive and supershift assays demonstrated binding sites for transcription factors Sp1, Sp3 (-408 to -413), and YY1 (-417 to -420). During differentiation of 3T3-L1 cells to adipocytes, the amount of Sp1 protein decreased by approximately 50% just prior to activation of PEMT. Transduction of 3T3-L1 adipocytes with retrovirus containing Sp1 cDNA demonstrated that Sp1 inhibited PEMT transcriptional activity. Similarly, short hairpin RNA directed against Sp1 in 3T3-L1 adipocytes enhanced PEMT transcriptional activation. Chromatin immunoprecipitation assays confirmed that Sp1 binds to the PEMT promoter, and this interaction decreases upon differentiation to adipocytes. These experiments directly link increased PEMT expression in adipocytes to decreased transcriptional expression of Sp1. In addition, our data established that Sp1 binding was required for tamoxifen-mediated inhibition of Pemt promoter activity.
Figures








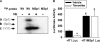
Similar articles
-
Transcriptional regulation of the hypoxia inducible factor-2alpha (HIF-2alpha) gene during adipose differentiation in 3T3-L1 cells.Biol Pharm Bull. 2006 Jan;29(1):49-54. doi: 10.1248/bpb.29.49. Biol Pharm Bull. 2006. PMID: 16394508
-
Mechanisms of divergent effects of activated peroxisome proliferator-activated receptor-γ on mitochondrial citrate carrier expression in 3T3-L1 fibroblasts and mature adipocytes.Biochim Biophys Acta. 2013 Jun;1831(6):1027-36. doi: 10.1016/j.bbalip.2013.01.014. Epub 2013 Jan 28. Biochim Biophys Acta. 2013. PMID: 23370576
-
Sp1 and Sp3 are involved in the full transcriptional activity of centromere protein H in human nasopharyngeal carcinoma cells.FEBS J. 2012 Aug;279(15):2714-26. doi: 10.1111/j.1742-4658.2012.08654.x. Epub 2012 Jun 25. FEBS J. 2012. PMID: 22682030
-
Physiological roles of phosphatidylethanolamine N-methyltransferase.Biochim Biophys Acta. 2013 Mar;1831(3):626-32. doi: 10.1016/j.bbalip.2012.07.017. Epub 2012 Jul 31. Biochim Biophys Acta. 2013. PMID: 22877991 Review.
-
Phospholipid methylation in mammals: from biochemistry to physiological function.Biochim Biophys Acta. 2014 Jun;1838(6):1477-87. doi: 10.1016/j.bbamem.2013.10.018. Epub 2013 Nov 1. Biochim Biophys Acta. 2014. PMID: 24184426 Review.
Cited by
-
Distinct infrastructure of lipid networks in visceral and subcutaneous adipose tissues in overweight humans.Am J Clin Nutr. 2020 Oct 1;112(4):979-990. doi: 10.1093/ajcn/nqaa195. Am J Clin Nutr. 2020. PMID: 32766878 Free PMC article.
-
Sequential synthesis and methylation of phosphatidylethanolamine promote lipid droplet biosynthesis and stability in tissue culture and in vivo.J Biol Chem. 2011 May 13;286(19):17338-50. doi: 10.1074/jbc.M111.234534. Epub 2011 Mar 22. J Biol Chem. 2011. PMID: 21454708 Free PMC article.
-
An organelle-specific photoactivation and dual-isotope labeling strategy reveals phosphatidylethanolamine metabolic flux.Chem Sci. 2023 Jan 16;14(7):1687-1695. doi: 10.1039/d2sc06069h. eCollection 2023 Feb 15. Chem Sci. 2023. PMID: 36819876 Free PMC article.
-
N-Myc and SP regulate phosphatidylserine synthase-1 expression in brain and glial cells.J Biol Chem. 2011 Jan 14;286(2):1061-73. doi: 10.1074/jbc.M110.158709. Epub 2010 Nov 10. J Biol Chem. 2011. PMID: 21068393 Free PMC article.
-
Phosphatidylethanolamine N-methyltransferase: from Functions to Diseases.Aging Dis. 2023 Jun 1;14(3):879-891. doi: 10.14336/AD.2022.1025. Aging Dis. 2023. PMID: 37191416 Free PMC article. Review.
References
-
- Vance D. E., Vance J. E. (eds) (2008) Biochemistry of Lipids, Lipoproteins and Membranes, 5th Ed., pp. 214–244, Elsevier B. V., San Diego, CA
-
- Vance D. E., Ridgway N. D. (1988) Prog. Lipid Res. 27, 61–79 - PubMed
-
- Bremer J., Greenberg D. M. (1960) Biochim. Biophys. Acta 37, 173–175 - PubMed
-
- Cui Z., Vance J. E., Chen M. H., Voelker D. R., Vance D. E. (1993) J. Biol. Chem. 268, 16655–16663 - PubMed
-
- Shields D. J., Lehner R., Agellon L. B., Vance D. E. (2003) J. Biol. Chem. 278, 2956–2962 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

