Overexpression of bacterial ethylene-forming enzyme gene in Trichoderma reesei enhanced the production of ethylene
- PMID: 20150979
- PMCID: PMC2820237
- DOI: 10.7150/ijbs.6.96
Overexpression of bacterial ethylene-forming enzyme gene in Trichoderma reesei enhanced the production of ethylene
Abstract
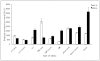
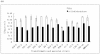
In order to efficiently utilize natural cellulose materials to produce ethylene, three expression vectors containing the ethylene-forming enzyme (efe) gene from Pseudomonas syringae pv. glycinea were constructed. The target gene was respectively controlled by different promoters: cbh I promoter from Trichoderma reesei cellobiohydrolases I gene, gpd promoter from Aspergillus nidulans glyceraldehyde-3-phosphate dehydrogenase gene and pgk I promoter from T. reesei 3-phosphoglycerate kinase I gene. After transforming into T. reesei QM9414, 43 stable transformants were obtained by PCR amplification and ethylene determination. Southern blot analysis of 14 transformants demonstrated that the efe gene was integrated into chromosomal DNA with copy numbers from 1 to 4. Reverse transcription polymerase chain reaction (RT-PCR) analysis of 6 transformants showed that the heterologous gene was transcribed. By using wheat straw as a carbon source, the ethylene production rates of aforementioned 14 transformants were measured. Transformant C30-3 with pgk I promoter had the highest ethylene production (4,012 nl h(-1) l(-1)). This indicates that agricultural wastes could be used to produce ethylene in recombinant filamentous fungus T. reesei.
Keywords: Trichoderma reesei; ethylene-forming enzyme; overexpression.; promoter; wheat straw.
Conflict of interest statement
Conflict of interests: The authors have declared that they have no conflict of interest exists.
Figures





Similar articles
-
Expression of ethylene-forming enzyme (EFE) of Pseudomonas syringae pv. glycinea in Trichoderma viride.Appl Microbiol Biotechnol. 2008 Sep;80(4):573-8. doi: 10.1007/s00253-008-1562-7. Epub 2008 Jun 25. Appl Microbiol Biotechnol. 2008. PMID: 18575855
-
Antisense inhibition of xylitol dehydrogenase gene, xdh1 from Trichoderma reesei.Lett Appl Microbiol. 2005;40(6):424-9. doi: 10.1111/j.1472-765X.2005.01685.x. Lett Appl Microbiol. 2005. PMID: 15892737
-
Improved biomass saccharification by Trichoderma reesei through heterologous expression of lacA gene from Trametes sp. AH28-2.J Biosci Bioeng. 2012 Jun;113(6):697-703. doi: 10.1016/j.jbiosc.2012.01.016. Epub 2012 Mar 3. J Biosci Bioeng. 2012. PMID: 22387233
-
Improved heterologous gene expression in Trichoderma reesei by cellobiohydrolase I gene (cbh1) promoter optimization.Acta Biochim Biophys Sin (Shanghai). 2008 Feb;40(2):158-65. doi: 10.1111/j.1745-7270.2008.00388.x. Acta Biochim Biophys Sin (Shanghai). 2008. PMID: 18235978
-
Construction of two vectors for gene expression in Trichoderma reesei.Plasmid. 2012 Jan;67(1):67-71. doi: 10.1016/j.plasmid.2011.10.002. Epub 2011 Oct 25. Plasmid. 2012. PMID: 22056690
Cited by
-
Ethylene synthesis and regulated expression of recombinant protein in Synechocystis sp. PCC 6803.PLoS One. 2012;7(11):e50470. doi: 10.1371/journal.pone.0050470. Epub 2012 Nov 21. PLoS One. 2012. PMID: 23185630 Free PMC article.
-
Metatranscriptomics and Amplicon Sequencing Reveal Mutualisms in Seagrass Microbiomes.Front Microbiol. 2018 Mar 15;9:388. doi: 10.3389/fmicb.2018.00388. eCollection 2018. Front Microbiol. 2018. PMID: 29599758 Free PMC article.
-
Cellulases and beyond: the first 70 years of the enzyme producer Trichoderma reesei.Microb Cell Fact. 2016 Jun 10;15(1):106. doi: 10.1186/s12934-016-0507-6. Microb Cell Fact. 2016. PMID: 27287427 Free PMC article. Review.
-
Identification and characterization of sugar-regulated promoters in Chaetomium thermophilum.BMC Biotechnol. 2023 Jul 8;23(1):19. doi: 10.1186/s12896-023-00791-9. BMC Biotechnol. 2023. PMID: 37422618 Free PMC article.
-
Ethylene production with engineered Synechocystis sp PCC 6803 strains.Microb Cell Fact. 2017 Feb 23;16(1):34. doi: 10.1186/s12934-017-0645-5. Microb Cell Fact. 2017. PMID: 28231787 Free PMC article.
References
-
- Hall MA, Smith AR. Ethylene and the responses of plants to stress. Bulgarian Journal Of Plant Physiology. 1995;21:71–79.
-
- Weingart H, Volksch B, Ullrich MS. Comparison of Ethylene Production by Pseudomonas syringae and Ralstonia solanacearum. Phytopathology. 1999;89:360–365. - PubMed
-
- Nagahama K, Ogawa T, Fujii T, Tazaki M, Tanase S, Morino Y, Fukuda H. Purification and properties of an ethylene-forming enzyme from Pseudomonas syringae pv. phaseolicola PK2. J Gen Microbiol. 1991;137:2281–2286. - PubMed
-
- Volksch B, Weingart H. Comparison of ethylene-producing Pseudomonas syringae strains isolated from kudzu (Pueraria lobata) with Pseudomonas syringae pv. phaseolicola and Pseudomonas syringae pv. glycinea. European Journal of Plant Pathology. 1997;103:795–802.
-
- Katsuya I, Masayoshi M, Yorinao I, Sumio T, Takahira O, Hideo F. Overexpression and in vitro reconstitution of the ethylene-forming enzyme from Pseudomonas syringae. Journal of Fermentation and Bioengineering. 1995;79:205–211.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

