Furfural induces reactive oxygen species accumulation and cellular damage in Saccharomyces cerevisiae
- PMID: 20150993
- PMCID: PMC2820483
- DOI: 10.1186/1754-6834-3-2
Furfural induces reactive oxygen species accumulation and cellular damage in Saccharomyces cerevisiae
Abstract
Background: Biofuels offer a viable alternative to petroleum-based fuel. However, current methods are not sufficient and the technology required in order to use lignocellulosic biomass as a fermentation substrate faces several challenges. One challenge is the need for a robust fermentative microorganism that can tolerate the inhibitors present during lignocellulosic fermentation. These inhibitors include the furan aldehyde, furfural, which is released as a byproduct of pentose dehydration during the weak acid pretreatment of lignocellulose. In order to survive in the presence of furfural, yeast cells need not only to reduce furfural to the less toxic furan methanol, but also to protect themselves and repair any damage caused by the furfural. Since furfural tolerance in yeast requires a functional pentose phosphate pathway (PPP), and the PPP is associated with reactive oxygen species (ROS) tolerance, we decided to investigate whether or not furfural induces ROS and its related cellular damage in yeast.
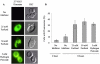
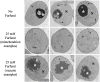
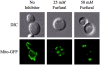
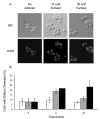
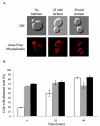
Results: We demonstrated that furfural induces the accumulation of ROS in Saccharomyces cerevisiae. In addition, furfural was shown to cause cellular damage that is consistent with ROS accumulation in cells which includes damage to mitochondria and vacuole membranes, the actin cytoskeleton and nuclear chromatin. The furfural-induced damage is less severe when yeast are grown in a furfural concentration (25 mM) that allows for eventual growth after an extended lag compared to a concentration of furfural (50 mM) that prevents growth.
Conclusion: These data suggest that when yeast cells encounter the inhibitor furfural, they not only need to reduce furfural into furan methanol but also to protect themselves from the cellular effects of furfural and repair any damage caused. The reduced cellular damage seen at 25 mM furfural compared to 50 mM furfural may be linked to the observation that at 25 mM furfural yeast were able to exit the furfural-induced lag phase and resume growth. Understanding the cellular effects of furfural will help direct future strain development to engineer strains capable of tolerating or remediating ROS and the effects of ROS.
Figures

References
-
- Bothast J, Saha B. Ethanol production from agricultural biomass substrates. Adv Appl Microbiol. 1997;44:261–286.
-
- Palmqvist E, Hahn-Hägerdal B. Fermentation of lignocellulosic hydrolysates. II: inhibitors and mechanisms of inhibition. Bioresour Technol. 2000;74:25–33. doi: 10.1016/S0960-8524(99)00161-3. - DOI
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

