Comparative study of the binding pockets of mammalian proprotein convertases and its implications for the design of specific small molecule inhibitors
- PMID: 20151049
- PMCID: PMC2820236
- DOI: 10.7150/ijbs.6.89
Comparative study of the binding pockets of mammalian proprotein convertases and its implications for the design of specific small molecule inhibitors
Abstract
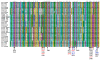
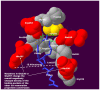
Proprotein convertases are enzymes that proteolytically cleave protein precursors in the secretory pathway to yield functional proteins. Seven mammalian subtilisin/Kex2p-like proprotein convertases have been identified: furin, PC1, PC2, PC4, PACE4, PC5 and PC7. The binding pockets of all seven proprotein convertases are evolutionarily conserved and highly similar. Among the seven proprotein convertases, the furin cleavage site motif has recently been characterized as a 20-residue motif that includes one core region P6-P2' inside the furin binding pocket. This study extended this information by examining the 3D structural environment of the furin binding pocket surrounding the core region P6-P2' of furin substrates. The physical properties of mutations in the binding pockets of the other six mammalian proprotein convertases were compared. The results suggest that: 1) mutations at two positions, Glu230 and Glu257, change the overall density of the negative charge of the binding pockets, and govern the substrate specificities of mammalian proprotein convertases; 2) two proprotein convertases (PC1 and PC2) may have reduced sensitivity for positively charged residues at substrate position P5 or P6, whereas the substrate specificities of three proprotein convertases (furin, PACE4, and PC5) are similar to each other. This finding led to a novel design of a short peptide pattern for small molecule inhibitors: [K/R]-X-V-X-K-R. Compared with the widely used small molecule dec-RVKR-cmk that inhibits all seven proprotein convertases, a finely-tuned derivative of the short peptide pattern [K/R]-X-V-X-K-R may have the potential to more effectively inhibit five of the proprotein convertases (furin, PC4, PACE4, PC5 and PC7) compared to the remaining two (PC1 and PC2). The results not only provide insights into the molecular evolution of enzyme function in the proprotein convertase family, but will also aid the study of the functional redundancy of proprotein convertases and the development of therapeutic applications.
Keywords: evolution of gene family.; mammalian proprotein convertases; small molecular inhibitor; substrate specificity.
Conflict of interest statement
Conflict of Interests: Sun Tian has had a full time job since PhD graduation in June 2007. Sun Tian and this project have been self-financed since then and this study was completed in the author's own time. The authors declare that no conflict of interest exists.
Figures

References
-
- Seidah NG, Chretien M. Proprotein and prohormone convertases: a family of subtilases generating diverse bioactive polypeptides. Brain Res. 1999;848:45–62. - PubMed
-
- Henrich S, Cameron A, Bourenkov GP, Kiefersauer R, Huber R, Lindberg I. et al. The crystal structure of the proprotein processing proteinase furin explains its stringent specificity. Nat Struct Biol. 2003;10:520–526. - PubMed
-
- Sun T. A 20 Residues Motif Delineates the Furin Cleavage Site and its Physical Properties May Influence Viral Fusion. Biochemistry Insights. 2009;2:9–20.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

