New functions for an old variant: no substitute for histone H3.3
- PMID: 20153629
- PMCID: PMC2860041
- DOI: 10.1016/j.gde.2010.01.003
New functions for an old variant: no substitute for histone H3.3
Abstract
Histone proteins often come in different variants serving specialized functions in addition to their fundamental role in packaging DNA. The metazoan histone H3.3 has been most closely associated with active transcription. Its role in histone replacement at active genes and promoters is conserved to the single histone H3 in yeast. However, recent genetic studies in flies have challenged its importance as a mark of active chromatin, and revealed unexpected insights into essential functions of H3.3 in the germline. With strikingly little amino acid sequence difference to the canonical H3, H3.3 therefore accomplishes a surprising variety of cellular and developmental processes.
2010 Elsevier Ltd. All rights reserved.
Figures

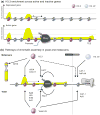

References
-
- Franklin SG, Zweidler A. Non-allelic variants of histones 2a, 2b and 3 in mammals. Nature. 1977;266:273–275. - PubMed
-
- Ahmad K, Henikoff S. The histone variant H3.3 marks active chromatin by replication-independent nucleosome assembly. Mol Cell. 2002;9:1191–1200. - PubMed
-
- Marzluff WF, Gongidi P, Woods KR, Jin J, Maltais LJ. The human and mouse replication-dependent histone genes. Genomics. 2002;80:487–498. - PubMed
-
- Rooney AP, Piontkivska H, Nei M. Molecular evolution of the nontandemly repeated genes of the histone 3 multigene family. Molecular Biology and Evolution. 2002;19:68–75. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

