Rapid purification of helicase proteins and in vitro analysis of helicase activity
- PMID: 20153831
- PMCID: PMC2892732
- DOI: 10.1016/j.ymeth.2010.02.008
Rapid purification of helicase proteins and in vitro analysis of helicase activity
Abstract
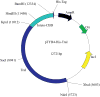
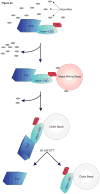
Most processes involving an organism's genetic material, including replication, repair and recombination, require access to single-stranded DNA as a template or reaction intermediate. To disrupt the hydrogen bonds between the two strands in double-stranded DNA, organisms utilize proteins called DNA helicases. DNA helicases use duplex DNA as a substrate to create single-stranded DNA in a reaction that requires ATP hydrolysis. Due to their critical role in cellular function, understanding the reaction catalyzed by helicases is essential to understanding DNA metabolism. Helicases are also important in many disease processes due to their role in DNA maintenance and replication. Here we discuss ways to rapidly purify helicases in sufficient quantity for biochemical analysis. We also briefly discuss potential substrates to use with helicases to establish some of their critical biochemical parameters. Through the use of methods that simplify the study of helicases, our understanding of these essential proteins can be accelerated.
Copyright (c) 2010 Elsevier Inc. All rights reserved.
Figures


Similar articles
-
Purification and characterization of two DNA helicases from calf thymus nuclei.J Biol Chem. 1991 Oct 25;266(30):20483-90. J Biol Chem. 1991. PMID: 1718963
-
DNA helicases: enzymes with essential roles in all aspects of DNA metabolism.Bioessays. 1994 Jan;16(1):13-22. doi: 10.1002/bies.950160103. Bioessays. 1994. PMID: 8141804 Review.
-
Escherichia coli DNA helicases: mechanisms of DNA unwinding.Mol Microbiol. 1992 Jan;6(1):5-14. doi: 10.1111/j.1365-2958.1992.tb00831.x. Mol Microbiol. 1992. PMID: 1310794 Review.
-
ATP hydrolysis stimulates binding and release of single stranded DNA from alternating subunits of the dimeric E. coli Rep helicase: implications for ATP-driven helicase translocation.J Mol Biol. 1996 Nov 1;263(3):411-22. doi: 10.1006/jmbi.1996.0585. J Mol Biol. 1996. PMID: 8918597
-
Escherichia coli DNA helicase I catalyzes a unidirectional and highly processive unwinding reaction.J Biol Chem. 1988 Mar 5;263(7):3208-15. J Biol Chem. 1988. PMID: 2830275
Cited by
-
Rapid, inexpensive, sequence-independent fluorescent labeling of phosphorothioate DNA.Biophys J. 2023 Apr 4;122(7):1211-1218. doi: 10.1016/j.bpj.2023.02.011. Epub 2023 Feb 14. Biophys J. 2023. PMID: 36793216 Free PMC article.
-
The UvrD303 hyper-helicase exhibits increased processivity.J Biol Chem. 2014 Jun 13;289(24):17100-10. doi: 10.1074/jbc.M114.565309. Epub 2014 May 5. J Biol Chem. 2014. PMID: 24798324 Free PMC article.
-
Insight into the biochemical mechanism of DNA helicases provided by bulk-phase and single-molecule assays.Methods. 2022 Aug;204:348-360. doi: 10.1016/j.ymeth.2021.12.002. Epub 2021 Dec 8. Methods. 2022. PMID: 34896247 Free PMC article. Review.
References
-
- Abdel-Monem M, Hoffmann-Berling H. Eur J Biochem. 1976;65:431–440. - PubMed
-
- Abdel-Monem M, Durwald H, Hoffmann-Berling H. Eur J Biochem. 1976;65:441–449. - PubMed
-
- Kuhn B, Abdel-Monem M, Krell H, Hoffmann-Berling H. J Biol Chem. 1979;254:11343–11350. - PubMed
-
- Matson SW, Bean DW, George JW. Bioessays. 1994;16:13–22. - PubMed
-
- Lohman TM, Bjornson KP. Annu Rev Biochem. 1996;65:169–214. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

