Genomic imprinting-an epigenetic gene-regulatory model
- PMID: 20153958
- PMCID: PMC2860637
- DOI: 10.1016/j.gde.2010.01.009
Genomic imprinting-an epigenetic gene-regulatory model
Abstract
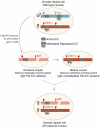
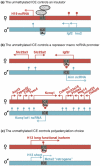
Epigenetic mechanisms (Box 1) are considered to play major gene-regulatory roles in development, differentiation and disease. However, the relative importance of epigenetics in defining the mammalian transcriptome in normal and disease states is unknown. The mammalian genome contains only a few model systems where epigenetic gene regulation has been shown to play a major role in transcriptional control. These model systems are important not only to investigate the biological function of known epigenetic modifications but also to identify new and unexpected epigenetic mechanisms in the mammalian genome. Here we review recent progress in understanding how epigenetic mechanisms control imprinted gene expression.
2010 Elsevier Ltd. All rights reserved.
Figures
References
-
- Carninci P. Is sequencing enlightenment ending the dark age of the transcriptome? Nat Methods. 2009;6:711–713. - PubMed
-
- Allis CD, Jenuwein T, Reinberg D. In: Epigenetics. edn 1. Marie-Laure Caparros AE, editor. Cold Spring Harbor Laboratory Press; New York: 2007.
-
-
Berger SL, Kouzarides T, Shiekhattar R, Shilatifard A. An operational definition of epigenetics. Genes Dev. 2009;23:781–783. The results of a meeting hosted by the Banbury Conference Center and Cold Spring Harbor Laboratory that discussed epigenetic control of genomic function reached a consensus definition of “epigenetics” to be considered by the broader community based on multiple mechanistic steps leading to the stable heritance of the epigenetic phenotype.
-
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

