Self-assembly of severe acute respiratory syndrome coronavirus membrane protein
- PMID: 20154085
- PMCID: PMC2857088
- DOI: 10.1074/jbc.M109.030270
Self-assembly of severe acute respiratory syndrome coronavirus membrane protein
Abstract
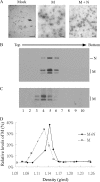
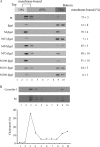
Coronavirus membrane (M) protein can form virus-like particles (VLPs) when coexpressed with nucleocapsid (N) or envelope (E) proteins, suggesting a pivotal role for M in virion assembly. Here we demonstrate the self-assembly and release of severe acute respiratory syndrome coronavirus (SARS-CoV) M protein in medium in the form of membrane-enveloped vesicles with densities lower than those of VLPs formed by M plus N. Although efficient N-N interactions require the presence of RNA, we found that M-M interactions were RNA-independent. SARS-CoV M was observed in both the Golgi area and plasma membranes of a variety of cells. Blocking M glycosylation does not appear to significantly affect M plasma membrane labeling intensity, M-containing vesicle release, or VLP formation. Results from a genetic analysis indicate involvement of the third transmembrane domain of M in plasma membrane-targeting signal. Fusion proteins containing M amino-terminal 50 residues encompassing the first transmembrane domain were found to be sufficient for membrane binding, multimerization, and Golgi retention. Surprisingly, we found that fusion proteins lacking all three transmembrane domains were still capable of membrane binding, Golgi retention, and interacting with M. The data suggest that multiple SARS-CoV M regions are involved in M self-assembly and subcellular localization.
Figures




Similar articles
-
The M, E, and N structural proteins of the severe acute respiratory syndrome coronavirus are required for efficient assembly, trafficking, and release of virus-like particles.J Virol. 2008 Nov;82(22):11318-30. doi: 10.1128/JVI.01052-08. Epub 2008 Aug 27. J Virol. 2008. PMID: 18753196 Free PMC article.
-
The SARS-CoV-2 envelope and membrane proteins modulate maturation and retention of the spike protein, allowing assembly of virus-like particles.J Biol Chem. 2021 Jan-Jun;296:100111. doi: 10.1074/jbc.RA120.016175. Epub 2020 Dec 3. J Biol Chem. 2021. PMID: 33229438 Free PMC article.
-
Characterization of severe acute respiratory syndrome coronavirus membrane protein.FEBS Lett. 2006 Feb 6;580(3):968-73. doi: 10.1016/j.febslet.2006.01.026. Epub 2006 Jan 19. FEBS Lett. 2006. PMID: 16442106 Free PMC article.
-
Properties of Coronavirus and SARS-CoV-2.Malays J Pathol. 2020 Apr;42(1):3-11. Malays J Pathol. 2020. PMID: 32342926 Review.
-
An overall picture of SARS coronavirus (SARS-CoV) genome-encoded major proteins: structures, functions and drug development.Curr Pharm Des. 2006;12(35):4539-53. doi: 10.2174/138161206779010459. Curr Pharm Des. 2006. PMID: 17168760 Review.
Cited by
-
Computational Study of the Ion and Water Permeation and Transport Mechanisms of the SARS-CoV-2 Pentameric E Protein Channel.Front Mol Biosci. 2020 Sep 23;7:565797. doi: 10.3389/fmolb.2020.565797. eCollection 2020. Front Mol Biosci. 2020. PMID: 33173781 Free PMC article.
-
The coronavirus E protein: assembly and beyond.Viruses. 2012 Mar;4(3):363-82. doi: 10.3390/v4030363. Epub 2012 Mar 8. Viruses. 2012. PMID: 22590676 Free PMC article. Review.
-
Membrane binding proteins of coronaviruses.Future Virol. 2019 Apr;14(4):275-286. doi: 10.2217/fvl-2018-0144. Epub 2019 Apr 29. Future Virol. 2019. PMID: 32201500 Free PMC article. Review.
-
Understanding the Molecular Biology of SARS-CoV-2 and the COVID-19 Pandemic: A Review.Infect Drug Resist. 2021 Jun 16;14:2259-2268. doi: 10.2147/IDR.S306441. eCollection 2021. Infect Drug Resist. 2021. PMID: 34163190 Free PMC article. Review.
-
Evaluation of Cell-Based and Surrogate SARS-CoV-2 Neutralization Assays.J Clin Microbiol. 2021 Sep 20;59(10):e0052721. doi: 10.1128/JCM.00527-21. Epub 2021 Jul 21. J Clin Microbiol. 2021. PMID: 34288726 Free PMC article.
References
-
- Lai M. M. (1987) Adv. Exp. Med. Biol. 218, 7–13 - PubMed
-
- Spaan W., Cavanagh D., Horzinek M. C. (1988) J. Gen. Virol. 69, 2939–2952 - PubMed
-
- Marra M. A., Jones S. J., Astell C. R., Holt R. A., Brooks-Wilson A., Butterfield Y. S., Khattra J., Asano J. K., Barber S. A., Chan S. Y., Cloutier A., Coughlin S. M., Freeman D., Girn N., Griffith O. L., Leach S. R., Mayo M., McDonald H., Montgomery S. B., Pandoh P. K., Petrescu A. S., Robertson A. G., Schein J. E., Siddiqui A., Smailus D. E., Stott J. M., Yang G. S., Plummer F., Andonov A., Artsob H., Bastien N., Bernard K., Booth T. F., Bowness D., Czub M., Drebot M., Fernando L., Flick R., Garbutt M., Gray M., Grolla A., Jones S., Feldmann H., Meyers A., Kabani A., Li Y., Normand S., Stroher U., Tipples G. A., Tyler S., Vogrig R., Ward D., Watson B., Brunham R. C., Krajden M., Petric M., Skowronski D. M., Upton C., Roper R. L. (2003) Science 300, 1399–1404 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

