Endogenous proteolytic cleavage of disease-associated prion protein to produce C2 fragments is strongly cell- and tissue-dependent
- PMID: 20154089
- PMCID: PMC2856230
- DOI: 10.1074/jbc.M109.083857
Endogenous proteolytic cleavage of disease-associated prion protein to produce C2 fragments is strongly cell- and tissue-dependent
Abstract
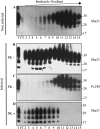
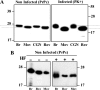
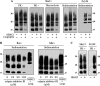
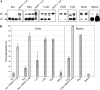
The abnormally folded form of the prion protein (PrP(Sc)) accumulating in nervous and lymphoid tissues of prion-infected individuals can be naturally cleaved to generate a N-terminal-truncated fragment called C2. Information about the identity of the cellular proteases involved in this process and its possible role in prion biology has remained limited and controversial. We investigated PrP(Sc) N-terminal trimming in different cell lines and primary cultured nerve cells, and in the brain and spleen tissue from transgenic mice infected by ovine and mouse prions. We found the following: (i) the full-length to C2 ratio varies considerably depending on the infected cell or tissue. Thus, in primary neurons and brain tissue, PrP(Sc) accumulated predominantly as untrimmed species, whereas efficient trimming occurred in Rov and MovS cells, and in spleen tissue. (ii) Although C2 is generally considered to be the counterpart of the PrP(Sc) proteinase K-resistant core, the N termini of the fragments cleaved in vivo and in vitro can actually differ, as evidenced by a different reactivity toward the Pc248 anti-octarepeat antibody. (iii) In lysosome-impaired cells, the ratio of full-length versus C2 species dramatically increased, yet efficient prion propagation could occur. Moreover, cathepsin but not calpain inhibitors markedly inhibited C2 formation, and in vitro cleavage by cathepsins B and L produced PrP(Sc) fragments lacking the Pc248 epitope, strongly arguing for the primary involvement of acidic hydrolases of the endolysosomal compartment. These findings have implications on the molecular analysis of PrP(Sc) and cell pathogenesis of prion infection.
Figures





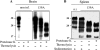
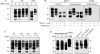

Similar articles
-
Calpain-dependent endoproteolytic cleavage of PrPSc modulates scrapie prion propagation.J Biol Chem. 2004 May 21;279(21):21948-56. doi: 10.1074/jbc.M400793200. Epub 2004 Mar 16. J Biol Chem. 2004. PMID: 15026410
-
Delayed progression of prion disease in mice by polyarginine-facilitated prevention of PrPSc propagation in the spleen.Neurotherapeutics. 2025 Apr;22(3):e00560. doi: 10.1016/j.neurot.2025.e00560. Epub 2025 Feb 26. Neurotherapeutics. 2025. PMID: 40011131 Free PMC article.
-
Modulation of proteinase K-resistant prion protein in cells and infectious brain homogenate by redox iron: implications for prion replication and disease pathogenesis.Mol Biol Cell. 2007 Sep;18(9):3302-12. doi: 10.1091/mbc.e07-04-0317. Epub 2007 Jun 13. Mol Biol Cell. 2007. PMID: 17567949 Free PMC article.
-
Evolving views in prion glycosylation: functional and pathological implications.Biochimie. 2003 Jan-Feb;85(1-2):33-45. doi: 10.1016/s0300-9084(03)00040-3. Biochimie. 2003. PMID: 12765773 Review.
-
Transgenic models of prion disease.Arch Virol Suppl. 2000;(16):113-24. doi: 10.1007/978-3-7091-6308-5_10. Arch Virol Suppl. 2000. PMID: 11214913 Review.
Cited by
-
Strain-specific role of RNAs in prion replication.J Virol. 2012 Oct;86(19):10494-504. doi: 10.1128/JVI.01286-12. Epub 2012 Jul 18. J Virol. 2012. PMID: 22811520 Free PMC article.
-
The strain-encoded relationship between PrP replication, stability and processing in neurons is predictive of the incubation period of disease.PLoS Pathog. 2011 Mar;7(3):e1001317. doi: 10.1371/journal.ppat.1001317. Epub 2011 Mar 17. PLoS Pathog. 2011. PMID: 21437239 Free PMC article.
-
MEK1 transduces the prion protein N2 fragment antioxidant effects.Cell Mol Life Sci. 2015 Apr;72(8):1613-29. doi: 10.1007/s00018-014-1777-y. Epub 2014 Nov 13. Cell Mol Life Sci. 2015. PMID: 25391659 Free PMC article.
-
Application of N-Terminal Labeling Methods Provide Novel Insights into Endoproteolysis of the Prion Protein in Vivo.ACS Chem Neurosci. 2024 Jan 3;15(1):134-146. doi: 10.1021/acschemneuro.3c00533. Epub 2023 Dec 14. ACS Chem Neurosci. 2024. PMID: 38095594 Free PMC article.
-
Interaction of prion protein with acetylcholinesterase: potential pathobiological implications in prion diseases.Acta Neuropathol Commun. 2015 Apr 3;3:18. doi: 10.1186/s40478-015-0188-0. Acta Neuropathol Commun. 2015. PMID: 25853328 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

