Real-time structural transitions are coupled to chemical steps in ATP hydrolysis by Eg5 kinesin
- PMID: 20154092
- PMCID: PMC2856982
- DOI: 10.1074/jbc.C110.103762
Real-time structural transitions are coupled to chemical steps in ATP hydrolysis by Eg5 kinesin
Abstract
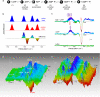
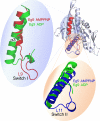
At the biochemical level, motor proteins are enzymatic molecules that function by converting chemical energy into mechanical motion. The key element for energy transduction and a major unresolved question common for all motor proteins is the coordination between the chemical and conformational steps in ATP hydrolysis. Here we show time-lapse monitoring of an in vitro ATP hydrolysis reaction by the motor domain of a human Kinesin-5 protein (Eg5) using difference Fourier transform infrared spectroscopy and UV photolysis of caged ATP. In this first continuous observation of a biological reaction coordinate from substrate to product, direct spectral markers for two catalytic events are measured: proton abstraction from nucleophilic water by the catalytic base and formation of the inorganic phosphate leaving group. Simultaneous examination of conformational switching in Eg5, in parallel with catalytic steps, shows structural transitions in solution consistent with published crystal structures of the prehydrolytic and ADP-bound states. In addition, we detect structural modifications in the Eg5 motor domain during bond cleavage between the beta- and gamma-phosphates of ATP. This conclusion challenges mechanochemical models for motor proteins that utilize only two stages of the catalytic cycle to drive force and motion.
Figures
Similar articles
-
ATP hydrolysis in Eg5 kinesin involves a catalytic two-water mechanism.J Biol Chem. 2010 Feb 19;285(8):5859-67. doi: 10.1074/jbc.M109.071233. Epub 2009 Dec 15. J Biol Chem. 2010. PMID: 20018897 Free PMC article.
-
Getting in sync with dimeric Eg5. Initiation and regulation of the processive run.J Biol Chem. 2008 Jan 25;283(4):2078-87. doi: 10.1074/jbc.M708354200. Epub 2007 Nov 25. J Biol Chem. 2008. PMID: 18037705 Free PMC article.
-
Disparity in allosteric interactions of monastrol with Eg5 in the presence of ADP and ATP: a difference FT-IR investigation.Biochemistry. 2004 Aug 10;43(31):9939-49. doi: 10.1021/bi048982y. Biochemistry. 2004. PMID: 15287721
-
Review: Mechanochemistry of the kinesin-1 ATPase.Biopolymers. 2016 Aug;105(8):476-82. doi: 10.1002/bip.22862. Biopolymers. 2016. PMID: 27120111 Free PMC article. Review.
-
Kinesin motors as molecular machines.Bioessays. 2003 Dec;25(12):1212-9. doi: 10.1002/bies.10358. Bioessays. 2003. PMID: 14635256 Review.
Cited by
-
The yeast kinesin-5 Cin8 interacts with the microtubule in a noncanonical manner.J Biol Chem. 2017 Sep 1;292(35):14680-14694. doi: 10.1074/jbc.M117.797662. Epub 2017 Jul 12. J Biol Chem. 2017. PMID: 28701465 Free PMC article.
-
Allosteric drug discrimination is coupled to mechanochemical changes in the kinesin-5 motor core.J Biol Chem. 2010 Jun 11;285(24):18650-61. doi: 10.1074/jbc.M109.092072. Epub 2010 Mar 18. J Biol Chem. 2010. PMID: 20299460 Free PMC article.
-
Common Mechanism of Activated Catalysis in P-loop Fold Nucleoside Triphosphatases-United in Diversity.Biomolecules. 2022 Sep 22;12(10):1346. doi: 10.3390/biom12101346. Biomolecules. 2022. PMID: 36291556 Free PMC article.
-
Kinesin-5: cross-bridging mechanism to targeted clinical therapy.Gene. 2013 Dec 1;531(2):133-49. doi: 10.1016/j.gene.2013.08.004. Epub 2013 Aug 14. Gene. 2013. PMID: 23954229 Free PMC article. Review.
-
Kinesin-5 allosteric inhibitors uncouple the dynamics of nucleotide, microtubule, and neck-linker binding sites.Biophys J. 2014 Nov 4;107(9):2204-13. doi: 10.1016/j.bpj.2014.09.019. Biophys J. 2014. PMID: 25418105 Free PMC article.
References
-
- Wojcik E. J., Dalrymple N. A., Alford S. R., Walker R. A., Kim S. (2004) Biochemistry 43, 9939–9949 - PubMed
-
- Du X., Frei H., Kim S. H. (2000) J. Biol. Chem. 275, 8492–8500 - PubMed
-
- Allin C., Gerwert K. (2001) Biochemistry 40, 3037–3046 - PubMed
-
- Liu M., Barth A. (2002) Biopolymers. 67, 267–270 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

