Distribution of virulence factors and molecular fingerprinting of Aeromonas species isolates from water and clinical samples: suggestive evidence of water-to-human transmission
- PMID: 20154106
- PMCID: PMC2849238
- DOI: 10.1128/AEM.02535-09
Distribution of virulence factors and molecular fingerprinting of Aeromonas species isolates from water and clinical samples: suggestive evidence of water-to-human transmission
Abstract
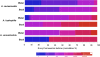
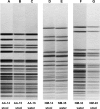
A total of 227 isolates of Aeromonas obtained from different geographical locations in the United States and different parts of the world, including 28 reference strains, were analyzed to determine the presence of various virulence factors. These isolates were also fingerprinted using biochemical identification and pulse-field gel electrophoresis (PFGE). Of these 227 isolates, 199 that were collected from water and clinical samples belonged to three major groups or complexes, namely, the A. hydrophila group, the A. caviae-A. media group, and the A. veronii-A. sobria group, based on biochemical profiles, and they had various pulsotypes. When virulence factor activities were examined, Aeromonas isolates obtained from clinical sources had higher cytotoxic activities than isolates obtained from water sources for all three Aeromonas species groups. Likewise, the production of quorum-sensing signaling molecules, such as N-acyl homoserine lactone, was greater in clinical isolates than in isolates from water for the A. caviae-A. media and A. hydrophila groups. Based on colony blot DNA hybridization, the heat-labile cytotonic enterotoxin gene and the DNA adenosine methyltransferase gene were more prevalent in clinical isolates than in water isolates for all three Aeromonas groups. Using colony blot DNA hybridization and PFGE, we obtained three sets of water and clinical isolates that had the same virulence signature and had indistinguishable PFGE patterns. In addition, all of these isolates belonged to the A. caviae-A. media group. The findings of the present study provide the first suggestive evidence of successful colonization and infection by particular strains of certain Aeromonas species after transmission from water to humans.
Figures

Similar articles
-
Identity, virulence genes, and clonal relatedness of Aeromonas isolates from patients with diarrhea and drinking water.Eur J Clin Microbiol Infect Dis. 2010 Sep;29(9):1163-72. doi: 10.1007/s10096-010-0982-3. Epub 2010 Jun 13. Eur J Clin Microbiol Infect Dis. 2010. PMID: 20549532
-
Identity and virulence properties of Aeromonas isolates from diseased fish, healthy controls and water environment in China.Lett Appl Microbiol. 2012 Sep;55(3):224-33. doi: 10.1111/j.1472-765X.2012.03281.x. Epub 2012 Jul 20. Lett Appl Microbiol. 2012. PMID: 22725694
-
Characterization of Aeromonas spp. isolated from humans with diarrhea, from healthy controls, and from surface water in Bangladesh.J Clin Microbiol. 1997 Feb;35(2):369-73. doi: 10.1128/jcm.35.2.369-373.1997. J Clin Microbiol. 1997. PMID: 9003598 Free PMC article.
-
Occurrence of potential virulence determinants in Aeromonas spp. isolated from different aquatic environments.J Appl Microbiol. 2023 Mar 1;134(3):lxad031. doi: 10.1093/jambio/lxad031. J Appl Microbiol. 2023. PMID: 36809788 Review.
-
A review on aeromoniasis in poultry: A bacterial disease of zoonotic nature.J Infect Dev Ctries. 2023 Jan 31;17(1):1-9. doi: 10.3855/jidc.17186. J Infect Dev Ctries. 2023. PMID: 36795920 Review.
Cited by
-
Precaution for using nucleic acid-based methods to detect Aeromonas.J Clin Microbiol. 2012 May;50(5):1829. doi: 10.1128/JCM.00077-12. Epub 2012 Feb 22. J Clin Microbiol. 2012. PMID: 22357502 Free PMC article. No abstract available.
-
Insight into the mobilome of Aeromonas strains.Front Microbiol. 2015 May 27;6:494. doi: 10.3389/fmicb.2015.00494. eCollection 2015. Front Microbiol. 2015. PMID: 26074893 Free PMC article. Review.
-
Classification of Aeromonas spp. isolated from water and clinical sources and distribution of virulence genes.Folia Microbiol (Praha). 2016 Nov;61(6):513-521. doi: 10.1007/s12223-016-0464-9. Epub 2016 Jul 14. Folia Microbiol (Praha). 2016. PMID: 27416863
-
Functional genomic characterization of virulence factors from necrotizing fasciitis-causing strains of Aeromonas hydrophila.Appl Environ Microbiol. 2014 Jul;80(14):4162-83. doi: 10.1128/AEM.00486-14. Epub 2014 May 2. Appl Environ Microbiol. 2014. PMID: 24795370 Free PMC article.
-
Anti-quorum Sensing and Protective Efficacies of Naringin Against Aeromonas hydrophila Infection in Danio rerio.Front Microbiol. 2020 Dec 3;11:600622. doi: 10.3389/fmicb.2020.600622. eCollection 2020. Front Microbiol. 2020. PMID: 33424802 Free PMC article.
References
-
- Abdullah, A. I., C. A. Hart, and C. Winstanley. 2003. Molecular characterization and distribution of virulence-associated genes amongst Aeromonas isolates from Libya. J. Appl. Microbiol. 95:1001-1007. - PubMed
-
- Abuhammour, W., R. A. Hasan, and D. Rogers. 2006. Necrotizing fasciitis caused by Aeromonas hydrophila in an immunocompetent child. Pediatr. Emerg. Care 22:48-51. - PubMed
-
- Aguilera-Arreola, M. G., C. Hernandez-Rodriguez, G. Zuniga, M. J. Figueras, and G. Castro-Escarpulli. 2005. Aeromonas hydrophila clinical and environmental ecotypes as revealed by genetic diversity and virulence genes. FEMS Microbiol. Lett. 242:231-240. - PubMed
-
- Albert, M. J., M. Ansaruzzaman, K. A. Talukder, A. K. Chopra, I. Kuhn, M. Rahman, A. S. Faruque, M. S. Islam, R. B. Sack, and R. Mollby. 2000. Prevalence of enterotoxin genes in Aeromonas spp. isolated from children with diarrhea, healthy controls, and the environment. J. Clin. Microbiol. 38:3785-3790. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

