Identification and analysis of the biosynthetic gene cluster encoding the thiopeptide antibiotic cyclothiazomycin in Streptomyces hygroscopicus 10-22
- PMID: 20154110
- PMCID: PMC2849233
- DOI: 10.1128/AEM.01790-09
Identification and analysis of the biosynthetic gene cluster encoding the thiopeptide antibiotic cyclothiazomycin in Streptomyces hygroscopicus 10-22
Abstract
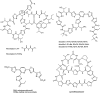
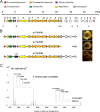
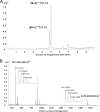
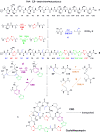
Thiopeptide antibiotics are an important class of natural products resulting from posttranslational modifications of ribosomally synthesized peptides. Cyclothiazomycin is a typical thiopeptide antibiotic that has a unique bridged macrocyclic structure derived from an 18-amino-acid structural peptide. Here we reported cloning, sequencing, and heterologous expression of the cyclothiazomycin biosynthetic gene cluster from Streptomyces hygroscopicus 10-22. Remarkably, successful heterologous expression of a 22.7-kb gene cluster in Streptomyces lividans 1326 suggested that there is a minimum set of 15 open reading frames that includes all of the functional genes required for cyclothiazomycin production. Six genes of these genes, cltBCDEFG flanking the structural gene cltA, were predicted to encode the enzymes required for the main framework of cyclothiazomycin, and two enzymes encoded by a putative operon, cltMN, were hypothesized to participate in the tailoring step to generate the tertiary thioether, leading to the final cyclization of the bridged macrocyclic structure. This rigorous bioinformatics analysis based on heterologous expression of cyclothiazomycin resulted in an ideal biosynthetic model for us to understand the biosynthesis of thiopeptides.
Figures

Similar articles
-
Regulation of the biosynthesis of thiopeptide antibiotic cyclothiazomycin by the transcriptional regulator SHJG8833 in Streptomyces hygroscopicus 5008.Microbiology (Reading). 2014 Jul;160(Pt 7):1379-1392. doi: 10.1099/mic.0.076901-0. Epub 2014 Apr 25. Microbiology (Reading). 2014. PMID: 24769844
-
Isolation and characterization of the gene cluster for biosynthesis of the thiopeptide antibiotic TP-1161.Appl Environ Microbiol. 2010 Nov;76(21):7093-101. doi: 10.1128/AEM.01442-10. Epub 2010 Sep 17. Appl Environ Microbiol. 2010. PMID: 20851988 Free PMC article.
-
Identification of the biosynthetic gene cluster for the antibiotic polyketide L-155,175 in Streptomyces hygroscopicus.Folia Microbiol (Praha). 2012 Nov;57(6):543-50. doi: 10.1007/s12223-012-0173-y. Epub 2012 Jun 6. Folia Microbiol (Praha). 2012. PMID: 22669556
-
Streptomycetes: Surrogate hosts for the genetic manipulation of biosynthetic gene clusters and production of natural products.Biotechnol Adv. 2019 Jan-Feb;37(1):1-20. doi: 10.1016/j.biotechadv.2018.10.003. Epub 2018 Oct 9. Biotechnol Adv. 2019. PMID: 30312648 Free PMC article. Review.
-
Antibiotics from microbes: converging to kill.Curr Opin Microbiol. 2009 Oct;12(5):520-7. doi: 10.1016/j.mib.2009.07.002. Epub 2009 Aug 18. Curr Opin Microbiol. 2009. PMID: 19695947 Free PMC article. Review.
Cited by
-
Sulfur-Containing Metabolites from Marine and Terrestrial Fungal Sources: Origin, Structures, and Bioactivities.Mar Drugs. 2022 Dec 7;20(12):765. doi: 10.3390/md20120765. Mar Drugs. 2022. PMID: 36547912 Free PMC article. Review.
-
Antimicrobial cyclic peptides for plant disease control.Plant Pathol J. 2015 Mar;31(1):1-11. doi: 10.5423/PPJ.RW.08.2014.0074. Epub 2015 Mar 31. Plant Pathol J. 2015. PMID: 25774105 Free PMC article.
-
New Scabimycins A-C Isolated from Streptomyces acidiscabies (Lu19992).Molecules. 2021 Sep 29;26(19):5922. doi: 10.3390/molecules26195922. Molecules. 2021. PMID: 34641466 Free PMC article.
-
Thiazolyl peptide antibiotic biosynthesis: a cascade of post-translational modifications on ribosomal nascent proteins.J Biol Chem. 2010 Sep 3;285(36):27525-31. doi: 10.1074/jbc.R110.135970. Epub 2010 Jun 3. J Biol Chem. 2010. PMID: 20522549 Free PMC article. Review.
-
Genomic and transcriptomic insights into the thermo-regulated biosynthesis of validamycin in Streptomyces hygroscopicus 5008.BMC Genomics. 2012 Jul 24;13:337. doi: 10.1186/1471-2164-13-337. BMC Genomics. 2012. PMID: 22827618 Free PMC article.
References
-
- Aoki, M., T. Ohtsuka, M. Yamada, Y. Ohba, H. Yoshizaki, H. Yasuno, T. Sano, J. Watanabe, K. Yokose, and H. Seto. 1991. Cyclothiazomycin, a novel polythiazole-containing peptide with renin inhibitory activity. Taxonomy, fermentation, isolation and physico-chemical characterization. J. Antibiot. (Tokyo) 44:582-588. - PubMed
-
- Bagley, M. C., J. W. Dale, E. A. Merritt, and X. Xiong. 2005. Thiopeptide antibiotics. Chem. Rev. 105:685-714. - PubMed
-
- Bagley, M. C., and X. Xiong. 2004. Stereoselective synthesis of the gamma-lactam hydrolysate of the thiopeptide cyclothiazomycin. Org. Lett. 6:3401-3404. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

