Di-adenosine tetraphosphate (Ap4A) metabolism impacts biofilm formation by Pseudomonas fluorescens via modulation of c-di-GMP-dependent pathways
- PMID: 20154123
- PMCID: PMC2901679
- DOI: 10.1128/JB.01571-09
Di-adenosine tetraphosphate (Ap4A) metabolism impacts biofilm formation by Pseudomonas fluorescens via modulation of c-di-GMP-dependent pathways
Abstract
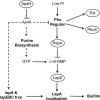
Dinucleoside tetraphosphates are common constituents of the cell and are thought to play diverse biological roles in organisms ranging from bacteria to humans. In this study we characterized two independent mechanisms by which di-adenosine tetraphosphate (Ap4A) metabolism impacts biofilm formation by Pseudomonas fluorescens. Null mutations in apaH, the gene encoding nucleoside tetraphosphate hydrolase, resulted in a marked increase in the cellular level of Ap4A. Concomitant with this increase, Pho regulon activation in low-inorganic-phosphate (P(i)) conditions was severely compromised. As a consequence, an apaH mutant was not sensitive to Pho regulon-dependent inhibition of biofilm formation. In addition, we characterized a Pho-independent role for Ap4A metabolism in regulation of biofilm formation. In P(i)-replete conditions Ap4A metabolism was found to impact expression and localization of LapA, the major adhesin regulating surface commitment by P. fluorescens. Increases in the level of c-di-GMP in the apaH mutant provided a likely explanation for increased localization of LapA to the outer membrane in response to elevated Ap4A concentrations. Increased levels of c-di-GMP in the apaH mutant were associated with increases in the level of GTP, suggesting that elevated levels of Ap4A may promote de novo purine biosynthesis. In support of this suggestion, supplementation with adenine could partially suppress the biofilm and c-di-GMP phenotypes of the apaH mutant. We hypothesize that changes in the substrate (GTP) concentration mediated by altered flux through nucleotide biosynthetic pathways may be a significant point of regulation for c-di-GMP biosynthesis and regulation of biofilm formation.
Figures




Similar articles
-
Phosphate-dependent modulation of c-di-GMP levels regulates Pseudomonas fluorescens Pf0-1 biofilm formation by controlling secretion of the adhesin LapA.Mol Microbiol. 2007 Feb;63(3):656-79. doi: 10.1111/j.1365-2958.2006.05539.x. Mol Microbiol. 2007. PMID: 17302799
-
From Input to Output: The Lap/c-di-GMP Biofilm Regulatory Circuit.Annu Rev Microbiol. 2020 Sep 8;74:607-631. doi: 10.1146/annurev-micro-011520-094214. Epub 2020 Jul 20. Annu Rev Microbiol. 2020. PMID: 32689917 Free PMC article. Review.
-
Systematic analysis of diguanylate cyclases that promote biofilm formation by Pseudomonas fluorescens Pf0-1.J Bacteriol. 2011 Sep;193(18):4685-98. doi: 10.1128/JB.05483-11. Epub 2011 Jul 15. J Bacteriol. 2011. PMID: 21764921 Free PMC article.
-
A c-di-GMP effector system controls cell adhesion by inside-out signaling and surface protein cleavage.PLoS Biol. 2011 Feb 1;9(2):e1000587. doi: 10.1371/journal.pbio.1000587. PLoS Biol. 2011. PMID: 21304920 Free PMC article.
-
Cyclic di-GMP: second messenger extraordinaire.Nat Rev Microbiol. 2017 May;15(5):271-284. doi: 10.1038/nrmicro.2016.190. Epub 2017 Feb 6. Nat Rev Microbiol. 2017. PMID: 28163311 Review.
Cited by
-
The structure of the CRISPR-associated protein Csa3 provides insight into the regulation of the CRISPR/Cas system.J Mol Biol. 2011 Jan 28;405(4):939-55. doi: 10.1016/j.jmb.2010.11.019. Epub 2010 Nov 18. J Mol Biol. 2011. PMID: 21093452 Free PMC article.
-
Re-evaluation of Diadenosine Tetraphosphate (Ap4A) From a Stress Metabolite to Bona Fide Secondary Messenger.Front Mol Biosci. 2020 Nov 17;7:606807. doi: 10.3389/fmolb.2020.606807. eCollection 2020. Front Mol Biosci. 2020. PMID: 33282915 Free PMC article. Review.
-
Pseudomonas aeruginosa: pathogenesis, virulence factors, antibiotic resistance, interaction with host, technology advances and emerging therapeutics.Signal Transduct Target Ther. 2022 Jun 25;7(1):199. doi: 10.1038/s41392-022-01056-1. Signal Transduct Target Ther. 2022. PMID: 35752612 Free PMC article. Review.
-
The regulator FleQ both transcriptionally and post-transcriptionally regulates the level of RTX adhesins of Pseudomonas fluorescens.J Bacteriol. 2023 Sep 26;205(9):e0015223. doi: 10.1128/jb.00152-23. Epub 2023 Sep 1. J Bacteriol. 2023. PMID: 37655913 Free PMC article.
-
Np4A alarmones function in bacteria as precursors to RNA caps.Proc Natl Acad Sci U S A. 2020 Feb 18;117(7):3560-3567. doi: 10.1073/pnas.1914229117. Epub 2020 Feb 4. Proc Natl Acad Sci U S A. 2020. PMID: 32019889 Free PMC article.
References
-
- Blanchin-Roland, S., S. Blanquet, J. M. Schmitter, and G. Fayat. 1986. The gene for Escherichia coli diadenosine tetraphosphatase is located immediately clockwise to folA and forms an operon with ksgA. Mol. Gen. Genet. 205:515-522. - PubMed
-
- Bochner, B. R., and B. N. Ames. 1982. Complete analysis of cellular nucleotides by two-dimensional thin layer chromatography. J. Biol. Chem. 257:9759-9769. - PubMed
-
- Bochner, B. R., P. C. Lee, S. W. Wilson, C. W. Cutler, and B. N. Ames. 1984. AppppA and related adenylylated nucleotides are synthesized as a consequence of oxidation stress. Cell 37:225-232. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

