Uncoupling of expression of an intronic microRNA and its myosin host gene by exon skipping
- PMID: 20154144
- PMCID: PMC2849460
- DOI: 10.1128/MCB.01370-09
Uncoupling of expression of an intronic microRNA and its myosin host gene by exon skipping
Abstract
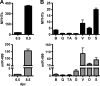
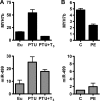
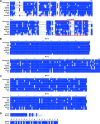
The ancient MYH7b gene, expressed in striated muscle and brain, encodes a sarcomeric myosin and the intronic microRNA miR-499. We find that skipping of an exon introduces a premature termination codon in the transcript that downregulates MYH7b protein production without affecting microRNA expression. Among other genes, endogenous miR-499 targets the 3' untranslated region of the transcription factor Sox6, which in turn acts as a repressor of MYH7b transcriptional activity. Thus, concerted transcription and alternative splicing uncouple the level of expression of MYH7b and miR-499 when their coexpression is not required.
Figures

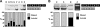
Similar articles
-
Functional divergence of the sarcomeric myosin, MYH7b, supports species-specific biological roles.J Biol Chem. 2023 Jan;299(1):102657. doi: 10.1016/j.jbc.2022.102657. Epub 2022 Nov 9. J Biol Chem. 2023. PMID: 36334627 Free PMC article.
-
Myosin 7b is a regulatory long noncoding RNA (lncMYH7b) in the human heart.J Biol Chem. 2021 Jan-Jun;296:100694. doi: 10.1016/j.jbc.2021.100694. Epub 2021 Apr 22. J Biol Chem. 2021. PMID: 33895132 Free PMC article.
-
Myh7b/miR-499 gene expression is transcriptionally regulated by MRFs and Eos.Nucleic Acids Res. 2012 Aug;40(15):7303-18. doi: 10.1093/nar/gks466. Epub 2012 May 25. Nucleic Acids Res. 2012. PMID: 22638570 Free PMC article.
-
The ancient sarcomeric myosins found in specialized muscles.Skelet Muscle. 2019 Mar 5;9(1):7. doi: 10.1186/s13395-019-0192-3. Skelet Muscle. 2019. PMID: 30836986 Free PMC article. Review.
-
Factors controlling cardiac myosin-isoform shift during hypertrophy and heart failure.J Mol Cell Cardiol. 2007 Oct;43(4):388-403. doi: 10.1016/j.yjmcc.2007.07.045. Epub 2007 Jul 21. J Mol Cell Cardiol. 2007. PMID: 17720186 Free PMC article. Review.
Cited by
-
Circulating miR-499a-5p Is a Potential Biomarker of MYH7-Associated Hypertrophic Cardiomyopathy.Int J Mol Sci. 2022 Mar 30;23(7):3791. doi: 10.3390/ijms23073791. Int J Mol Sci. 2022. PMID: 35409153 Free PMC article.
-
miR-499 protects cardiomyocytes from H 2O 2-induced apoptosis via its effects on Pdcd4 and Pacs2.RNA Biol. 2014;11(4):339-50. doi: 10.4161/rna.28300. Epub 2014 Feb 27. RNA Biol. 2014. PMID: 24646523 Free PMC article.
-
Cholino-ncRNAs modulate sex-specific- and age-related acetylcholine signals.FEBS Lett. 2020 Jul;594(14):2185-2198. doi: 10.1002/1873-3468.13789. Epub 2020 May 4. FEBS Lett. 2020. PMID: 32330292 Free PMC article. Review.
-
Myricetin improves endurance capacity by inducing muscle fiber type conversion via miR-499.Nutr Metab (Lond). 2019 May 2;16:27. doi: 10.1186/s12986-019-0353-8. eCollection 2019. Nutr Metab (Lond). 2019. PMID: 31073320 Free PMC article.
-
Differential regulation of MCM7 and its intronic miRNA cluster miR-106b-25 during megakaryopoiesis induced polyploidy.RNA Biol. 2014;11(9):1137-47. doi: 10.4161/rna.36136. RNA Biol. 2014. PMID: 25483046 Free PMC article.
References
-
- Allen, D. L., B. C. Harrison, A. Maass, M. L. Bell, W. C. Byrnes, and L. A. Leinwand. 2001. Cardiac and skeletal muscle adaptations to voluntary wheel running in the mouse. J. Appl. Physiol. 90:1900-1908. - PubMed
-
- Antos, C. L., T. A. McKinsey, M. Dreitz, L. M. Hollingsworth, C. L. Zhang, K. Schreiber, H. Rindt, R. J. Gorczynski, and E. N. Olson. 2003. Dose-dependent blockade to cardiomyocyte hypertrophy by histone deacetylase inhibitors. J. Biol. Chem. 278:28930-28937. - PubMed
-
- Aznarez, I., Y. Barash, O. Shai, D. He, J. Zielenski, L. C. Tsui, J. Parkinson, B. J. Frey, J. M. Rommens, and B. J. Blencowe. 2008. A systematic analysis of intronic sequences downstream of 5′ splice sites reveals a widespread role for U-rich motifs and TIA1/TIAL1 proteins in alternative splicing regulation. Genome Res. 18:1247-1258. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
