The AAA+ ATPase ATAD3A controls mitochondrial dynamics at the interface of the inner and outer membranes
- PMID: 20154147
- PMCID: PMC2849464
- DOI: 10.1128/MCB.00007-10
The AAA+ ATPase ATAD3A controls mitochondrial dynamics at the interface of the inner and outer membranes
Abstract
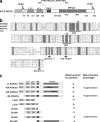
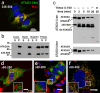
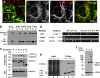
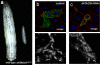
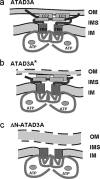
Dynamic interactions between components of the outer (OM) and inner (IM) membranes control a number of critical mitochondrial functions such as channeling of metabolites and coordinated fission and fusion. We identify here the mitochondrial AAA(+) ATPase protein ATAD3A specific to multicellular eukaryotes as a participant in these interactions. The N-terminal domain interacts with the OM. A central transmembrane segment (TMS) anchors the protein in the IM and positions the C-terminal AAA(+) ATPase domain in the matrix. Invalidation studies in Drosophila and in a human steroidogenic cell line showed that ATAD3A is required for normal cell growth and cholesterol channeling at contact sites. Using dominant-negative mutants, including a defective ATP-binding mutant and a truncated 50-amino-acid N-terminus mutant, we showed that ATAD3A regulates dynamic interactions between the mitochondrial OM and IM sensed by the cell fission machinery. The capacity of ATAD3A to impact essential mitochondrial functions and organization suggests that it possesses unique properties in regulating mitochondrial dynamics and cellular functions in multicellular organisms.
Figures


Similar articles
-
ATPase-deficient mitochondrial inner membrane protein ATAD3A disturbs mitochondrial dynamics in dominant hereditary spastic paraplegia.Hum Mol Genet. 2017 Apr 15;26(8):1432-1443. doi: 10.1093/hmg/ddx042. Hum Mol Genet. 2017. PMID: 28158749 Free PMC article.
-
The calcium-dependent interaction between S100B and the mitochondrial AAA ATPase ATAD3A and the role of this complex in the cytoplasmic processing of ATAD3A.Mol Cell Biol. 2010 Jun;30(11):2724-36. doi: 10.1128/MCB.01468-09. Epub 2010 Mar 29. Mol Cell Biol. 2010. PMID: 20351179 Free PMC article.
-
"ATAD3C regulates ATAD3A assembly and function in the mitochondrial membrane".Free Radic Biol Med. 2024 Feb 1;211:114-126. doi: 10.1016/j.freeradbiomed.2023.12.006. Epub 2023 Dec 12. Free Radic Biol Med. 2024. PMID: 38092275
-
Emerging Links between Control of Mitochondrial Protein ATAD3A and Cancer.Int J Mol Sci. 2020 Oct 25;21(21):7917. doi: 10.3390/ijms21217917. Int J Mol Sci. 2020. PMID: 33113782 Free PMC article. Review.
-
ATAD3A on the Path to Cancer.Adv Exp Med Biol. 2019;1134:259-269. doi: 10.1007/978-3-030-12668-1_14. Adv Exp Med Biol. 2019. PMID: 30919342 Review.
Cited by
-
A subcellular map of the human kinome.Elife. 2021 May 14;10:e64943. doi: 10.7554/eLife.64943. Elife. 2021. PMID: 33988507 Free PMC article.
-
A splice variant in ATAD3A expands the clinical and genetic spectrum of Harel-Yoon syndrome.Neurol Genet. 2020 Jun 3;6(4):e452. doi: 10.1212/NXG.0000000000000452. eCollection 2020 Aug. Neurol Genet. 2020. PMID: 32607449 Free PMC article. No abstract available.
-
Loss of mitochondrial ATPase ATAD3A contributes to nonalcoholic fatty liver disease through accumulation of lipids and damaged mitochondria.J Biol Chem. 2022 Jun;298(6):102008. doi: 10.1016/j.jbc.2022.102008. Epub 2022 May 2. J Biol Chem. 2022. PMID: 35513069 Free PMC article.
-
ATAD3A has a scaffolding role regulating mitochondria inner membrane structure and protein assembly.Cell Rep. 2021 Dec 21;37(12):110139. doi: 10.1016/j.celrep.2021.110139. Cell Rep. 2021. PMID: 34936866 Free PMC article.
-
ATAD3A-related pontocerebellar hypoplasia: new patients and insights into phenotypic variability.Orphanet J Rare Dis. 2023 Apr 24;18(1):92. doi: 10.1186/s13023-023-02689-3. Orphanet J Rare Dis. 2023. PMID: 37095554 Free PMC article.
References
-
- Bird, I. M., N. A. Hanley, R. A. Word, J. M. Mathis, J. L. McCarthy, J. I. Mason, and W. E. Rainey. 1993. Human NCI-H295 adrenocortical carcinoma cells: a model for angiotensin-II-responsive aldosterone secretion. Endocrinology 133:1555-1561. - PubMed
-
- Bogenhagen, D. F., D. Rousseau, and S. Burke. 2008. The layered structure of human mitochondrial DNA nucleoids. J. Biol. Chem. 283:3665-3675. - PubMed
-
- Boldogh, I. R., and L. A. Pon. 2007. Mitochondria on the move. Trends Cell Biol. 17:502-510. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
