STAT3 is constitutively phosphorylated on serine 727 residues, binds DNA, and activates transcription in CLL cells
- PMID: 20154216
- PMCID: PMC2918366
- DOI: 10.1182/blood-2009-10-230060
STAT3 is constitutively phosphorylated on serine 727 residues, binds DNA, and activates transcription in CLL cells
Abstract
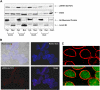
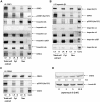
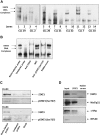
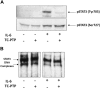
Chronic lymphocytic leukemia (CLL) is the most common leukemia in the Western hemisphere, but its pathogenesis is still poorly understood. Constitutive tyrosine phosphorylation (p) of signal transducer and activator of transcription (STAT) 3 occurs in several solid tumors and hematologic malignancies. In CLL, however, STAT3 is constitutively phosphorylated on serine 727, not tyrosine 705, residues. Because the biologic significance of serine pSTAT3 in CLL is not known, we studied peripheral blood cells of 106 patients with CLL and found that, although tyrosine pSTAT3 was inducible, serine pSTAT3 was constitutive in all patients studied, regardless of blood count, disease stage, or treatment status. In addition, we demonstrated that constitutive serine pSTAT3 translocates to the nucleus by the karyopherin-beta nucleocytoplasmic system and binds DNA. Dephosphorylation of inducible tyrosine pSTAT3 did not affect STAT3-DNA binding, suggesting that constitutive serine pSTAT3 binds DNA. Furthermore, infection of CLL cells with lentiviral STAT3-small hairpin RNA reduced the expression of several STAT3-regulated survival and proliferation genes and induced apoptosis, suggesting that constitutive serine pSTAT3 initiates transcription in CLL cells. Taken together, our data suggest that constitutive phosphorylation of STAT3 on serine 727 residues is a hallmark of CLL and that STAT3 be considered a therapeutic target in this disease.
Figures

Comment in
-
Lentiviral vectors and transduction of human cancer B cells.Blood. 2010 Jul 22;116(3):498-500; author reply 500. doi: 10.1182/blood-2010-03-276014. Blood. 2010. PMID: 20651085 No abstract available.
Similar articles
-
B lymphocytes from patients with chronic lymphocytic leukemia contain signal transducer and activator of transcription (STAT) 1 and STAT3 constitutively phosphorylated on serine residues.J Clin Invest. 1997 Dec 15;100(12):3140-8. doi: 10.1172/JCI119869. J Clin Invest. 1997. PMID: 9399961 Free PMC article.
-
Constitutive Phosphorylation of STAT3 by the CK2-BLNK-CD5 Complex.Mol Cancer Res. 2017 May;15(5):610-618. doi: 10.1158/1541-7786.MCR-16-0291. Epub 2017 Jan 27. Mol Cancer Res. 2017. PMID: 28130399 Free PMC article.
-
Viability and stress protection of chronic lymphoid leukemia cells involves overactivation of mitochondrial phosphoSTAT3Ser727.Cell Death Dis. 2014 Oct 9;5(10):e1451. doi: 10.1038/cddis.2014.393. Cell Death Dis. 2014. PMID: 25299776 Free PMC article.
-
The Important Role of STAT3 in Chronic Lymphocytic Leukaemia Biology.Klin Onkol. 2020 Winter;33(1):32-38. doi: 10.14735/amko202032. Klin Onkol. 2020. PMID: 32075387 Review. English.
-
Importance of STAT3 signalling in cancer, metastasis and therapeutic interventions.Cell Signal. 2022 Apr;92:110275. doi: 10.1016/j.cellsig.2022.110275. Epub 2022 Feb 3. Cell Signal. 2022. PMID: 35122990 Review.
Cited by
-
Two naturally occurring terpenes, dehydrocostuslactone and costunolide, decrease intracellular GSH content and inhibit STAT3 activation.PLoS One. 2011;6(5):e20174. doi: 10.1371/journal.pone.0020174. Epub 2011 May 18. PLoS One. 2011. PMID: 21625597 Free PMC article.
-
Clinico-Biological Implications of Modified Levels of Cytokines in Chronic Lymphocytic Leukemia: A Possible Therapeutic Role.Cancers (Basel). 2020 Feb 24;12(2):524. doi: 10.3390/cancers12020524. Cancers (Basel). 2020. PMID: 32102441 Free PMC article. Review.
-
Stat3 binds to mtDNA and regulates mitochondrial gene expression in keratinocytes.J Invest Dermatol. 2014 Jul;134(7):1971-1980. doi: 10.1038/jid.2014.68. Epub 2014 Feb 4. J Invest Dermatol. 2014. PMID: 24496235 Free PMC article.
-
Role of IL-9 and STATs in hematological malignancies (Review).Oncol Lett. 2014 Mar;7(3):602-610. doi: 10.3892/ol.2013.1761. Epub 2013 Dec 16. Oncol Lett. 2014. PMID: 24520283 Free PMC article.
-
Prognostic and Therapeutic Value of Apolipoprotein A and a New Risk Scoring System Based on Apolipoprotein A and Adenosine Deaminase in Chronic Lymphocytic Leukemia.Front Oncol. 2021 Jul 1;11:698572. doi: 10.3389/fonc.2021.698572. eCollection 2021. Front Oncol. 2021. PMID: 34277446 Free PMC article.
References
-
- Yee KW, O'Brien SM. Chronic lymphocytic leukemia: diagnosis and treatment. Mayo Clin Proc. 2006;81(8):1105–1129. - PubMed
-
- Chiorazzi N, Rai KR, Ferrarini M. Chronic lymphocytic leukemia. N Engl J Med. 2005;352(8):804–815. - PubMed
-
- Bueso-Ramos CE, Ferrajoli A, Medeiros LJ, Keating MJ, Estrov Z. Aberrant morphology, proliferation, and apoptosis of B-cell chronic lymphocytic leukemia cells. Hematology. 2004;9(4):279–286. - PubMed
-
- Calin GA, Croce CM. Genomics of chronic lymphocytic leukemia microRNAs as new players with clinical significance. Semin Oncol. 2006;33(2):167–173. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

