Common variants at 7p21 are associated with frontotemporal lobar degeneration with TDP-43 inclusions
- PMID: 20154673
- PMCID: PMC2828525
- DOI: 10.1038/ng.536
Common variants at 7p21 are associated with frontotemporal lobar degeneration with TDP-43 inclusions
Abstract
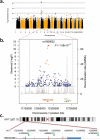
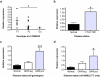
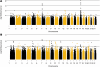
Frontotemporal lobar degeneration (FTLD) is the second most common cause of presenile dementia. The predominant neuropathology is FTLD with TAR DNA-binding protein (TDP-43) inclusions (FTLD-TDP). FTLD-TDP is frequently familial, resulting from mutations in GRN (which encodes progranulin). We assembled an international collaboration to identify susceptibility loci for FTLD-TDP through a genome-wide association study of 515 individuals with FTLD-TDP. We found that FTLD-TDP associates with multiple SNPs mapping to a single linkage disequilibrium block on 7p21 that contains TMEM106B. Three SNPs retained genome-wide significance following Bonferroni correction (top SNP rs1990622, P = 1.08 x 10(-11); odds ratio, minor allele (C) 0.61, 95% CI 0.53-0.71). The association replicated in 89 FTLD-TDP cases (rs1990622; P = 2 x 10(-4)). TMEM106B variants may confer risk of FTLD-TDP by increasing TMEM106B expression. TMEM106B variants also contribute to genetic risk for FTLD-TDP in individuals with mutations in GRN. Our data implicate variants in TMEM106B as a strong risk factor for FTLD-TDP, suggesting an underlying pathogenic mechanism.
Figures
Comment in
-
Deciphering genetic susceptibility to frontotemporal lobar dementia.Nat Genet. 2010 Mar;42(3):189-90. doi: 10.1038/ng0310-189. Nat Genet. 2010. PMID: 20179730 No abstract available.
References
-
- Neary D, et al. Frontotemporal lobar degeneration: a consensus on clinical diagnostic criteria. Neurology. 1998;51:1546–54. - PubMed
-
- McKhann GM, et al. Clinical and pathological diagnosis of frontotemporal dementia: report of the Work Group on Frontotemporal Dementia and Pick's Disease. Arch Neurol. 2001;58:1803–9. - PubMed
-
- Mercy L, Hodges JR, Dawson K, Barker RA, Brayne C. Incidence of early-onset dementias in Cambridgeshire, United Kingdom. Neurology. 2008;71:1496–9. - PubMed
-
- Ratnavalli E, Brayne C, Dawson K, Hodges JR. The prevalence of frontotemporal dementia. Neurology. 2002;58:1615–21. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
- AG19610/AG/NIA NIH HHS/United States
- AG08671/AG/NIA NIH HHS/United States
- P50 NS053488/NS/NINDS NIH HHS/United States
- AG005136/AG/NIA NIH HHS/United States
- NS53488/NS/NINDS NIH HHS/United States
- AG05134/AG/NIA NIH HHS/United States
- NS044233/NS/NINDS NIH HHS/United States
- P30 AG013854/AG/NIA NIH HHS/United States
- R01 NS044266/NS/NINDS NIH HHS/United States
- 75480/CAPMC/ CIHR/Canada
- P30 AG010124/AG/NIA NIH HHS/United States
- 089701/WT_/Wellcome Trust/United Kingdom
- P30 AG028377/AG/NIA NIH HHS/United States
- P50 AG008671/AG/NIA NIH HHS/United States
- AG05146/AG/NIA NIH HHS/United States
- R01 AG015116/AG/NIA NIH HHS/United States
- P50 AG005142/AG/NIA NIH HHS/United States
- AG16573/AG/NIA NIH HHS/United States
- AG05138/AG/NIA NIH HHS/United States
- AG10124/AG/NIA NIH HHS/United States
- P50 AG005131/AG/NIA NIH HHS/United States
- P30 AG010133/AG/NIA NIH HHS/United States
- AG005681/AG/NIA NIH HHS/United States
- NS44266/NS/NINDS NIH HHS/United States
- P50 AG016574/AG/NIA NIH HHS/United States
- P50 AG005146/AG/NIA NIH HHS/United States
- P01 AG017586/AG/NIA NIH HHS/United States
- NS15655/NS/NINDS NIH HHS/United States
- AG15116/AG/NIA NIH HHS/United States
- AG12300/AG/NIA NIH HHS/United States
- P50 AG008702/AG/NIA NIH HHS/United States
- AG03991/AG/NIA NIH HHS/United States
- P01 AG003991/AG/NIA NIH HHS/United States
- AG005131/AG/NIA NIH HHS/United States
- P50 AG005681/AG/NIA NIH HHS/United States
- P30 AG013846/AG/NIA NIH HHS/United States
- AG16570/AG/NIA NIH HHS/United States
- UL1 RR025741/RR/NCRR NIH HHS/United States
- P50 AG005136/AG/NIA NIH HHS/United States
- P30 AG012300/AG/NIA NIH HHS/United States
- NS038372/NS/NINDS NIH HHS/United States
- AG13846/AG/NIA NIH HHS/United States
- P50 AG016573/AG/NIA NIH HHS/United States
- AG02219/AG/NIA NIH HHS/United States
- P01 AG019724/AG/NIA NIH HHS/United States
- P50 AG016570/AG/NIA NIH HHS/United States
- P50 AG005134/AG/NIA NIH HHS/United States
- P30 AG008017/AG/NIA NIH HHS/United States
- P30 AG010161/AG/NIA NIH HHS/United States
- P01 NS015655/NS/NINDS NIH HHS/United States
- R01 AG018440/AG/NIA NIH HHS/United States
- AG05133/AG/NIA NIH HHS/United States
- G0701441/MRC_/Medical Research Council/United Kingdom
- AG008017/AG/NIA NIH HHS/United States
- G0301152/MRC_/Medical Research Council/United Kingdom
- AG025688/AG/NIA NIH HHS/United States
- P50 AG025688/AG/NIA NIH HHS/United States
- AG17586/AG/NIA NIH HHS/United States
- R37 AG018440/AG/NIA NIH HHS/United States
- P50 AG005133/AG/NIA NIH HHS/United States
- AG010133/AG/NIA NIH HHS/United States
- P01 AG002219/AG/NIA NIH HHS/United States
- P50 NS038372/NS/NINDS NIH HHS/United States
- AG033101/AG/NIA NIH HHS/United States
- ImNIH/Intramural NIH HHS/United States
- P50 AG005138/AG/NIA NIH HHS/United States
- AG18440/AG/NIA NIH HHS/United States
- AG16582/AG/NIA NIH HHS/United States
- R01 NS065782/NS/NINDS NIH HHS/United States
- AG10161/AG/NIA NIH HHS/United States
- AG13854/AG/NIA NIH HHS/United States
- P01 NS044233/NS/NINDS NIH HHS/United States
- AG08702/AG/NIA NIH HHS/United States
- P01 AG003949/AG/NIA NIH HHS/United States
- P30 AG010129/AG/NIA NIH HHS/United States
- P30 AG019610/AG/NIA NIH HHS/United States
- P50 AG016582/AG/NIA NIH HHS/United States
- AG16574/AG/NIA NIH HHS/United States
- AG010129/AG/NIA NIH HHS/United States
- AG05142/AG/NIA NIH HHS/United States
- AG028377/AG/NIA NIH HHS/United States
- G0600676/MRC_/Medical Research Council/United Kingdom
- AG03949/AG/NIA NIH HHS/United States
- AG5131/AG/NIA NIH HHS/United States
- G9724461/MRC_/Medical Research Council/United Kingdom
- G0601943/MRC_/Medical Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

