TGF-beta-mediated phosphorylation of hnRNP E1 induces EMT via transcript-selective translational induction of Dab2 and ILEI
- PMID: 20154680
- PMCID: PMC2830561
- DOI: 10.1038/ncb2029
TGF-beta-mediated phosphorylation of hnRNP E1 induces EMT via transcript-selective translational induction of Dab2 and ILEI
Abstract
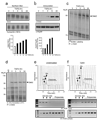
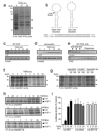
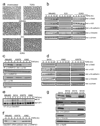
Transforming growth factor-beta (TGF-beta) induces epithelial-mesenchymal transdifferentiation (EMT) accompanied by cellular differentiation and migration. Despite extensive transcriptomic profiling, the identification of TGF-beta-inducible, EMT-specific genes has met with limited success. Here we identify a post-transcriptional pathway by which TGF-beta modulates the expression of EMT-specific proteins and of EMT itself. We show that heterogeneous nuclear ribonucleoprotein E1 (hnRNP E1) binds a structural, 33-nucleotide TGF-beta-activated translation (BAT) element in the 3' untranslated region of disabled-2 (Dab2) and interleukin-like EMT inducer (ILEI) transcripts, and represses their translation. TGF-beta activation leads to phosphorylation at Ser 43 of hnRNP E1 by protein kinase Bbeta/Akt2, inducing its release from the BAT element and translational activation of Dab2 and ILEI messenger RNAs. Modulation of hnRNP E1 expression or its post-translational modification alters the TGF-beta-mediated reversal of translational silencing of the target transcripts and EMT. These results suggest the existence of a TGF-beta-inducible post-transcriptional regulon that controls EMT during the development and metastatic progression of tumours.
Figures


References
-
- Bierie B, Moses HL. TGF-beta and cancer. Cytokine Growth Factor Rev. 2006;17:29–40. - PubMed
-
- Derynck R, Akhurst RJ, Balmain A. TGF-beta signaling in tumor suppression and cancer progression. Nature Genet. 2001;29:117–129. - PubMed
-
- Zavadil J, Bottinger EP. TGF-beta and epithelial-to-mesenchymal transitions. Oncogene. 2005;24:5764–5774. - PubMed
-
- Thiery JP, Sleeman JP. Complex networks orchestrate epithelial-mesenchymal transitions. Nature Rev. Mol. Cell. Biol. 2006;7:131–142. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

