Validation of reference genes for gene expression analysis in chicory (Cichorium intybus) using quantitative real-time PCR
- PMID: 20156357
- PMCID: PMC2830926
- DOI: 10.1186/1471-2199-11-15
Validation of reference genes for gene expression analysis in chicory (Cichorium intybus) using quantitative real-time PCR
Abstract
Background: Quantitative real-time reverse transcriptase polymerase chain reaction (qRT-PCR) is a sensitive technique for quantifying gene expression levels. One or more appropriate reference genes must be selected to accurately compare mRNA transcripts across different samples and tissues. Thus far, only actin-2 has been used as a reference gene for qRT-PCR in chicory, and a full comparison of several candidate reference genes in chicory has not yet been reported.
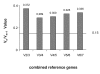
Results: Seven candidate reference genes, including nicotinamide adenine dinucleotide dehydrogenase (NADHD), actin (ACT), beta-tubulin (TUB), glyceraldehyde-3-phosphate-dehydrogenase (GADPH), histone H3 (H3), elongation factor 1-alpha (EF) and 18S rRNA (rRNA) were selected to study the expression stability for normalisation of gene expression in chicory. Primer specificity and amplification efficiency were verified for each gene. The expression stability of these genes was analysed across chicory root and leaf tissues using geNorm, NormFinder and BestKeeper software. ACT, EF, and rRNA were the most stable genes as identified by the three different analysis methods. In addition, the use of ACT, EF and GAPDH as reference genes was illustrated by analysing 1-FEHII (FEHII) expression in chicory root and leaf tissues. These analyses revealed the biological variation in FEHII transcript expression among the tissues studied, and between individual plants.
Conclusions: geNorm, NormFinder, and BestKeeper analyses indicated that ACT, EF and rRNA had the highest expression stability across leaf and root tissues, while GAPDH and NADHD showed relatively low expression stability. The results of this study emphasise the importance of validating reference genes for qRT-PCR analysis in chicory. The use of the most stable reference genes such as ACT and EF allows accurate normalisation of gene expression in chicory leaf and root tissues.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

