Identification of antisense RNA stem-loops that inhibit RNA-protein interactions using a bacterial reporter system
- PMID: 20156995
- PMCID: PMC2879510
- DOI: 10.1093/nar/gkq027
Identification of antisense RNA stem-loops that inhibit RNA-protein interactions using a bacterial reporter system
Abstract
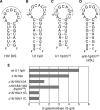
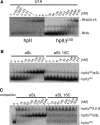
Many well-characterized examples of antisense RNAs from prokaryotic systems involve hybridization of the looped regions of stem-loop RNAs, presumably due to the high thermodynamic stability of the resulting loop-loop and loop-linear interactions. In this study, the identification of RNA stem-loops that inhibit U1A protein binding to the hpII RNA through RNA-RNA interactions was attempted using a bacterial reporter system based on phage lambda N-mediated antitermination. As a result, loop sequences possessing 7-8 base complementarity to the 5' region of the boxA element important for functional antitermination complex formation, but not the U1 hpII loop, were identified. In vitro and in vivo mutational analysis strongly suggested that the selected loop sequences were binding to the boxA region, and that the structure of the antisense stem-loop was important for optimal inhibitory activity. Next, in an attempt to demonstrate the ability to inhibit the interaction between the U1A protein and the hpII RNA, the rational design of an RNA stem-loop that inhibits U1A-binding to a modified hpII was carried out. Moderate inhibitory activity was observed, showing that it is possible to design and select antisense RNA stem-loops that disrupt various types of RNA-protein interactions.
Figures



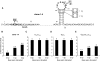

Similar articles
-
Identification of antisense RNA stem-loops that inhibit RNA-protein interactions using a bacterial reporter system.Nucleic Acids Symp Ser (Oxf). 2006;(50):77-8. doi: 10.1093/nass/nrl038. Nucleic Acids Symp Ser (Oxf). 2006. PMID: 17150825
-
Two structurally different RNA molecules are bound by the spliceosomal protein U1A using the same recognition strategy.Structure. 1996 May 15;4(5):621-31. doi: 10.1016/s0969-2126(96)00066-4. Structure. 1996. PMID: 8736559
-
Molecular dynamics simulations of the complex between human U1A protein and hairpin II of U1 small nuclear RNA and of free RNA in solution.Biophys J. 1999 Sep;77(3):1284-305. doi: 10.1016/S0006-3495(99)76979-1. Biophys J. 1999. PMID: 10465742 Free PMC article.
-
RNA-protein interactions. Diverse modes of recognition.Curr Biol. 1995 Mar 1;5(3):249-51. doi: 10.1016/s0960-9822(95)00051-0. Curr Biol. 1995. PMID: 7540101 Review.
-
Model of the interaction between the UI A small nuclear ribonucleoprotein and UI small nuclear RNA stem/loop II.Biochem Soc Trans. 1993 Aug;21 ( Pt 3)(3):605-9. doi: 10.1042/bst0210605. Biochem Soc Trans. 1993. PMID: 8224475 Review. No abstract available.
References
-
- Held DM, Kissel JD, Patterson JT, Nickens DG, Burke DH. HIV-1 inactivation by nucleic acid aptamers. Front Biosci. 2006;11:89–112. - PubMed
-
- Thomas JR, Hergenrother PJ. Targeting RNA with small molecules. Chem. Rev. 2008;108:1171–1224. - PubMed
-
- Crooke ST. Molecular mechanisms of action of antisense drugs. Biochim. Biophys. Acta. 1999;1489:31–44. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

